Kir3.3
Description: potassium inwardly-rectifying channel, subfamily J, member 9 Gene: Kcnj9 Alias: Kir3.3, Girk3, Kcnj9
KCNJ9 (also known as GIRK3; KIR3.3) encodes the integral membrane protein Kir3.3. This is a potassium inwardly-rectifying channel, subfamily J, member 9, which has a greater tendency to allow potassium to flow into a cell rather than out of a cell, is controlled by G-proteins. It associates with another G-protein-activated potassium channel to form a heteromultimeric pore-forming complex.
http://www.ncbi.nlm.nih.gov/gene/3765
G protein-gated inwardly rectifying potassium (GIRK or Kir3) channel activity is important for regulating excitability in the heart and brain (Stanfield et al. 2002 [957]).
Experimental data
Rat Kir3.3 gene in CHO host cells |
||
|
Click for details 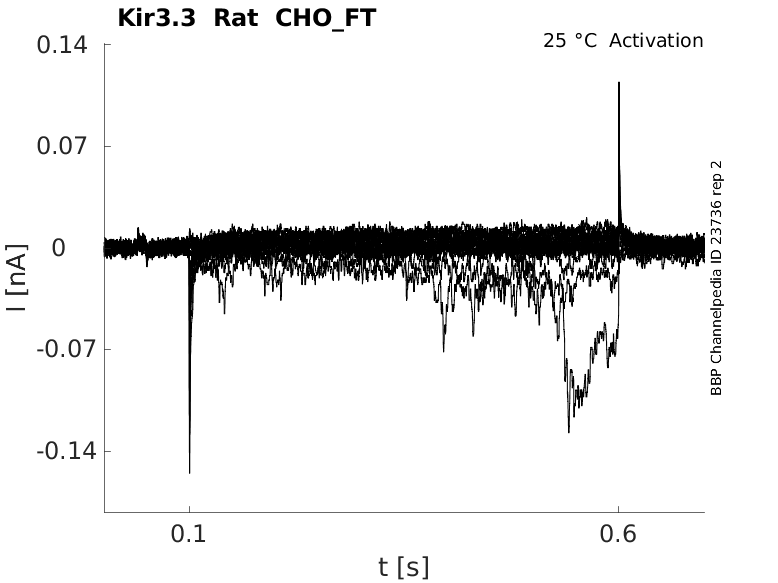
25 °Cshow 37 cells |
Click for details 
35 °Cshow 45 cells |
|
Gene
Transcript
| Species | NCBI accession | Length (nt) | |
|---|---|---|---|
| Human | NM_004983.3 | 4202 | |
| Mouse | NM_001360808.1 | 3914 | |
| Rat | NM_053834.2 | 2308 |
Protein Isoforms
Isoforms
Post-Translational Modifications
Structure
Kir3.3 predicted AlphaFold size
Methodology for AlphaFold size prediction and disclaimer are available here
Single channel Conductance of Kir3.3/Kir3.1 in CHO

Kir3.3 is expressed in mouse brain. Kofuji [195]
5-HT autoreceptors and G protein-gated inwardly rectifying potassium channels (Kir3/GIRK family), as well as their functional connectivity, has been demonstrated in raphe neurons (Penington [994]).
Kir3.3 subunit protein is expressed in raphe-derived axons at the light and electron microscopic level, but none of the other Kir3 subfamily members or the KATP channel subunits Kir6.1 and Kir6.2. (Pruess [993])
The GluR7 receptor gene GRIK3 is located on chromosome 1p34–33, where a significant linkage with schizo- phrenia has been reported (DeLisi et al., 2002 [991]. Significant changes of GluR7 expression in schizophrenia have been reported in multiple brain regions (Sokolov, 1998 [992]). (From [989])
The serotonergic system of the brainstem raphe is involved in mood control, the sleep-wake cycle, auto- nomic function, and stress response. The axons of certain dorsal raphe neurons form a dense serotonergic supraependymal plexus lining the brain ventricles, likely regulating ependymal metabolism and activity including ciliary movements and glucose homeostasis. In raphe neurons, serotonin exerts its function partly via 5-HT autoreceptors and G protein-gated inwardly rectifying potassium channels (Kir3/GIRK). Kir3.3 containing potassium channels may be of functional importance in autoregulation and excitability of supraependymal fibres and the complex serotonergic regulation along the parenchyma/CSF border. (Pruess [993])
Recombinantly expressed NCAM180 specifically reduces inward currents of neuron-specific Kir3.1/ 3.2 and Kir3.1/3.3 but not Kir3.1/3.4 channels in Xenopus oocytes and CHO cells (Delling [990]).
NCAM, TrkB and Kir3.3 interact directly with each other via their intracellular domains. Overexpression of the developmentally late appearing Kir3.3 subunit leads to a decrease in NCAM-mediated neurite outgrowth. Kir3.3 chan- nel expression at the cell surface; thus activity is regulated by NCAM and TrkB independently of BDNF ligand binding. These observations indicate that the interplay of recognition molecules, neurotrophin receptors, and ion channels regulate neurite outgrowth. Kleene [988]
G-Protein
Kir3 channels are activated following stimulation of G protein-coupled receptors (GPCRs) that use the Gi/o family of G proteins. Stimulation of the GPCR promotes exchange of GDP for GTP on the Gα subunit which, in turn, leads to activation of the Gα subunit and the Gβγ dimer. Gβγ dimers bind to and activate Kir3 channels (Reuveny et al. 1994 [968]; Wickman et al. 1994 [969]; Huang et al. 1995 [970]). Gα subunits are required for terminating Kir3 activation. The intrinsic GTPase activity of the Gα subunit hydrolyses GTP, leading to inactivation of the Gβγ dimer. Regulator of G protein signalling (RGS) proteins accelerate the GTPase activity of Gα subunits (GAP), leading to faster activation and deactivation of Kir3 channels (Doupnik et al. 1997 [971]). (From Fowler [965])
References
Kofuji P
et al.
Evidence that neuronal G-protein-gated inwardly rectifying K+ channels are activated by G beta gamma subunits and function as heteromultimers.
Proc. Natl. Acad. Sci. U.S.A.,
1995
Jul
3
, 92 (6542-6).
Stanfield PR
et al.
Constitutively active and G-protein coupled inward rectifier K+ channels: Kir2.0 and Kir3.0.
Rev. Physiol. Biochem. Pharmacol.,
2002
, 145 (47-179).
Fowler CE
et al.
Evidence for association of GABA(B) receptors with Kir3 channels and regulators of G protein signalling (RGS4) proteins.
J. Physiol. (Lond.),
2007
Apr
1
, 580 (51-65).
Reuveny E
et al.
Activation of the cloned muscarinic potassium channel by G protein beta gamma subunits.
Nature,
1994
Jul
14
, 370 (143-6).
Wickman KD
et al.
Recombinant G-protein beta gamma-subunits activate the muscarinic-gated atrial potassium channel.
Nature,
1994
Mar
17
, 368 (255-7).
Huang CL
et al.
Evidence that direct binding of G beta gamma to the GIRK1 G protein-gated inwardly rectifying K+ channel is important for channel activation.
Neuron,
1995
Nov
, 15 (1133-43).
Doupnik CA
et al.
RGS proteins reconstitute the rapid gating kinetics of gbetagamma-activated inwardly rectifying K+ channels.
Proc. Natl. Acad. Sci. U.S.A.,
1997
Sep
16
, 94 (10461-6).
Kleene R
et al.
Functional consequences of the interactions between the neural cell adhesion molecule NCAM, the receptor tyrosine kinase TrkB and the inwardly rectifying K+ channel Kir3.3.
,
2010
Jul
6
, ().
Kilic G
et al.
Are GRIK3 (T928G) gene variants in schizophrenia patients different from those in their first-degree relatives?
Psychiatry Res,
2010
Jan
30
, 175 (43-6).
Delling M
et al.
The neural cell adhesion molecule regulates cell-surface delivery of G-protein-activated inwardly rectifying potassium channels via lipid rafts.
J. Neurosci.,
2002
Aug
15
, 22 (7154-64).
DeLisi LE
et al.
Genome-wide scan for linkage to schizophrenia in a Spanish-origin cohort from Costa Rica.
Am. J. Med. Genet.,
2002
Jul
8
, 114 (497-508).
Sokolov BP
Expression of NMDAR1, GluR1, GluR7, and KA1 glutamate receptor mRNAs is decreased in frontal cortex of "neuroleptic-free" schizophrenics: evidence on reversible up-regulation by typical neuroleptics.
J. Neurochem.,
1998
Dec
, 71 (2454-64).
Prüss H
et al.
Expression of Kir3.3 potassium channel subunits in supraependymal axons.
Neurosci. Lett.,
2008
Nov
7
, 445 (89-93).
Penington NJ
et al.
Whole-cell recordings of inwardly rectifying K+ currents activated by 5-HT1A receptors on dorsal raphe neurones of the adult rat.
J. Physiol. (Lond.),
1993
Sep
, 469 (387-405).
Credits
To cite this page: [Contributors] Channelpedia https://channelpedia.epfl.ch/wikipages/48/ , accessed on 2026 Feb 23