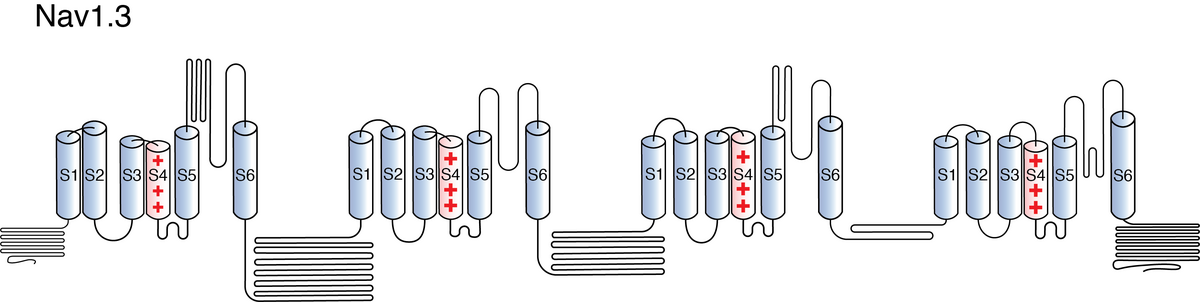
Nav1.3
Description: sodium channel, voltage-gated, type III, alpha Gene: Scn3a Alias: nav1.3, scn3a
Nav1.3, encoded by the gene scn3a, is a sodium, voltage-gated, type 3, alpha subunit channel. Nav1.3 is primarily expressed in the CNS during early development. It is responsible for the initiation of action potential. Mutations to the channel are often linked to neuropathic pain disorder.
Experimental data
Mouse Nav1.3 gene in CHO host cells |
||
|
Click for details 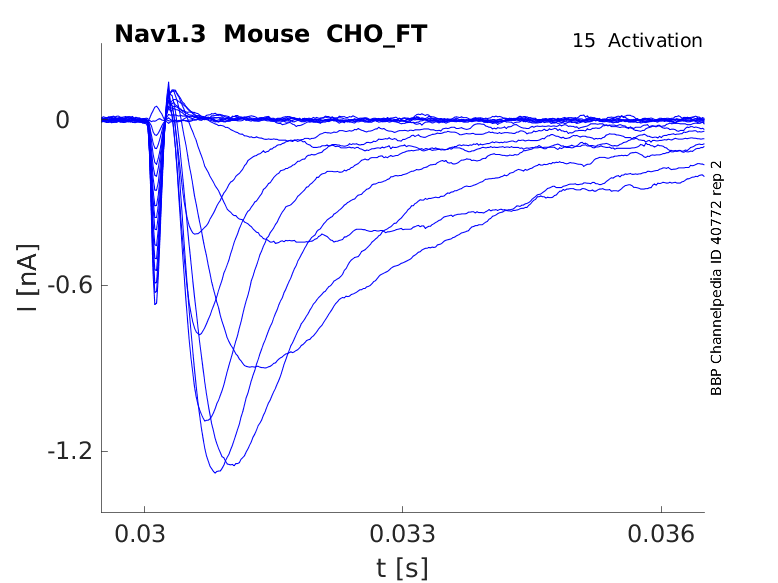
15 °Cshow 26 cells |
Click for details 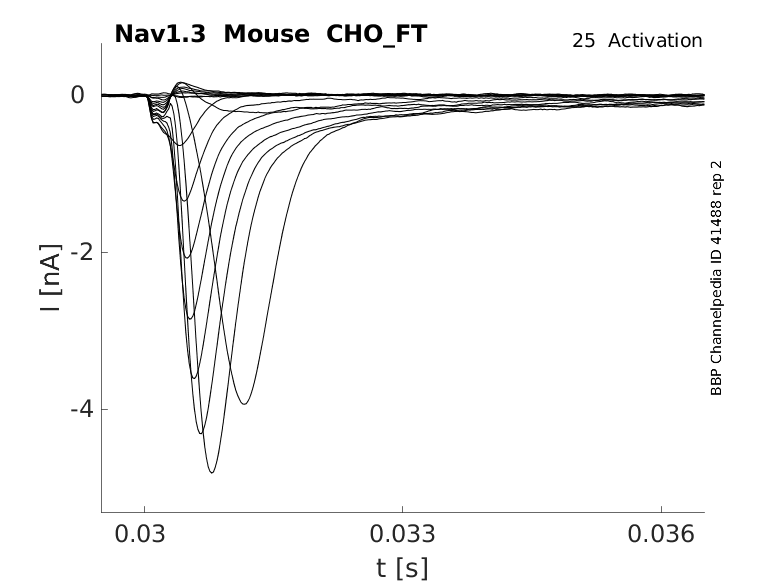
25 °Cshow 40 cells |
Click for details 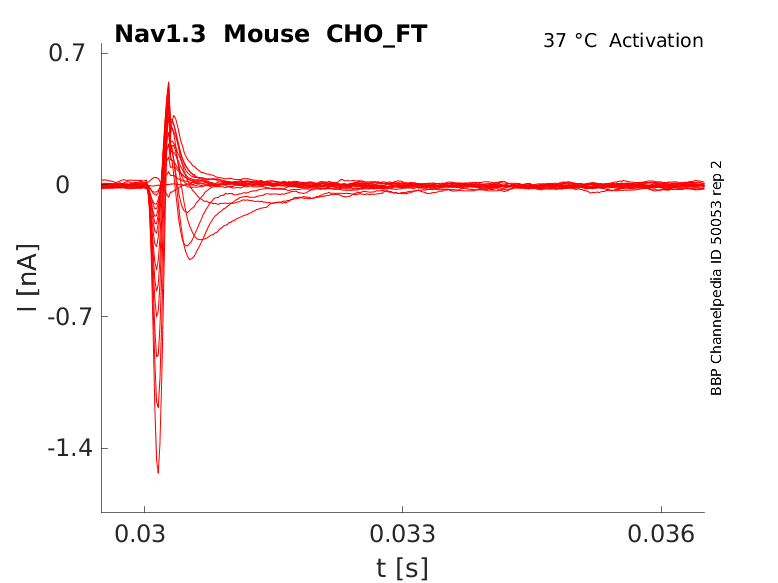
35 °Cshow 36 cells |
Nav1.3 is encoded by the gene scn3a present at the position 2q24.3 in the human genome. The length of scn3a gene is up of 28 exons, 26 of which are coding and exons 1 and 2 being non-coding. [2112]
scn3a is present in the same cluster as the genes coding for the other voltage gated sodium channels scn1a (nav1.1) and scn2a (nav1.2) [2120]
Multiple alternative splice scn3a variants have been observed (see table below) [2112]:
The best studied scn3a splice variants are referred to as the adult and neonatal forms. They result from the alternative mRNA splicing of either exon 5A (adult) or 5N (neonatal). Both variants have almost identical coding regions, with only a 21 nucleotide difference. The expression of either variant seems to be mutually exclusive and the selection of these two exons are developmentally regulated (see Expression & Distribution) [2113]
There is also the alternative splicing of Exon 12, which encodes a region of the DI-DII intracellular loop containing important phosphorylation sites that modulate channel kinetics.
| Species | NCBI accession | Length (nt) | |
|---|---|---|---|
| Human | NM_006922.4 | 9102 | |
| Mouse | NM_001355166.1 | 9707 | |
| Rat | NM_013119.2 | 6822 |
The human Nav1.3 protein is composed of 2000 amino acid (aa) and has a molecular weight of 227 Kda.
There exists a number of protein isoforms that arise from the translation of the aforementioned transcript variants:
- Nav1.3A and Nav1.3N isoforms are nearly identical in sequences with only a single amino acid difference at position 209 , specifying either aspartic acid (IIIA) or serine (IIIN). [2291]
- The alternative splicing event at exon 12 generates 4 Nav1.3 isoforms (12v1, 12v2, 12v3, 12v4) in humans and 3 isoforms (NaCh III, NaCh IIIa, NaCh IIIb) in rats [41]. The functional consequence of this splicing event are unknown
Isoforms
Like most mammalian proteins, Nav1.3 is subject to a series of post translational modifications (PTM).
Glycosylation is crucial for the normal activity of the Nav1.3. Deglycosylation experiments have been shown to change Nav1.3 kinetics, leading to a higher activation threshold and slower inactivation of the ion channel [2121].
Nav1.3 is potentially ubiquitinated as it contains a conserved PY motif in its COOH-terminal sequence known to interact with known to bind to a protein-ubiquitin ligase of the Nedd4 family. This PTM could promote its endocytosis of the protein [2122].
Visual Representation of Nav1.3 Structure
Methodology for visual representation of structure available here
Like all voltage gated sodium channels, Nav1.3 is made up of a single protein comprised of 4 homologous domains (DI-DIV). Each domain is made up of 6 transmembrane subunits. S1-4 form the voltage sensing domain (VSD) whereas the S5-6 form the pore module (PM). The S4 subunit of each domain contains a series of positively charged residues. When membrane depolarization occurs, these charged residues cause the movement of the S4 subunit, inducing a conformational change in S5-S6, opening of the channel and allowing the entry of sodium ions into the cell. Soon after opening, rapid inactivation of Nav1.3 is instigated by the binding of the IFM motif, found in the loop between D3 and D4, to a hydrophobic receptor site next to the S6 in D4. This binding causes the shift of S6, allosterically closing the channel, thus deactivating the channel [2115]. Nav1.3 then returns to its resting state following the hyperpolarization of the cell membrane [2116].
The structure of human Nav1.3, in complex with Navβ1/Navβ2, was resolved via cryo-electron microscopy, giving us a detailed insight to the structural features of Nav1.3. The Navβ1 subunit was shown to bind to the α-subunit through extensive interactions between the N-terminal domain and Domain I extracellular loop, as well as packing of the C-terminal helix against Domain III subunit 2. The β2 subunit was also shown to interact via disulfide bonds between C55 (β2) and C911 (α). [2290]
The approximative size/surface of Nav1.3 can be determined via the resolved or predicted structures.
Nav1.3 predicted AlphaFold size
Methodology for AlphaFold size prediction and disclaimer are available here
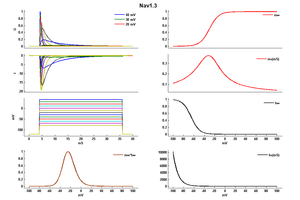
The Nav1.3 channel demonstrates rapid kinetics in terms of activation and inactivation, accompanied by a fast recovery from the inactivated state. The recovery from inactivation occurs more swiftly at negative potentials, while displaying slower dynamics at positive potentials. Notably, the progression of closed-state inactivation occurs gradually, resulting in the generation of prominent ramp currents during gradual depolarization. The relatively rapid recovery from inactivation and the slow closed-state inactivation kinetics of Nav1.3 channels suggest the potential for neurons expressing Nav1.3 to possess a lower activation threshold and/or exhibit a higher frequency of firing. [43]
Single Channel Unitary Conductance
Single channel unitary conductance is determined experimentally.
For Nav1.3, single channel unitary conductance has yet to be determined experimentally.
Model
A single kinetic model for all human voltage-gated sodium channels (Balbi et al, 2017)
https://modeldb.science/230137
Species : Human | Gene: scn3a
Host cell: HEK293 | Temperature: RT (to 25 C by Q10)
Formalism: Markov | States: C1, C2, O1, O2, I1, I2
Implementation: NEURON | Simulation: Nav13_a.mod
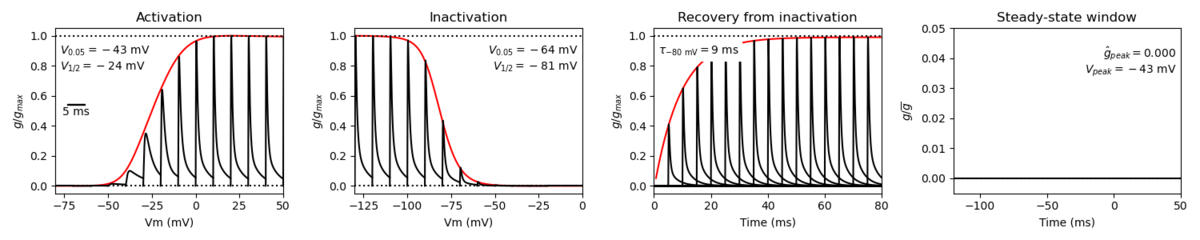
Membrane Systems Group 2023
Species: Mouse | Gene: Scn3a
Host cell: CHO | Temperature: 25 C
Formalism: Hodgkin-Huxley | Gates: m3, h
Implementation: NEURON | Simulation: Nav13__mCHO25c.mod
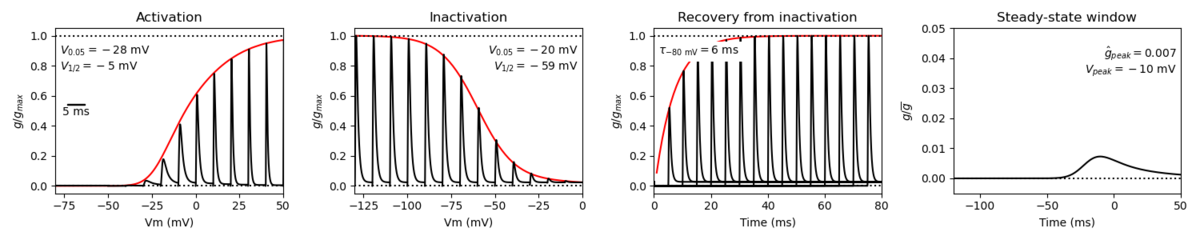
Model Nav1.3 (ID=43)
| Animal | rat | |
| CellType | Neocortical | |
| Age | 0 Days | |
| Temperature | 23.0°C | |
| Reversal | 50.0 mV | |
| Ion | Na + | |
| Ligand ion | ||
| Reference | [43] T R Cummins et. al; J. Neurosci. 2001 Aug 15 | |
| mpower | 3.0 | |
| m Alpha | (0.182 * ((v)- -26))/(1-(exp(-((v)- -26)/9))) If v neq -26 | |
| m Beta | (0.124 * (-(v) -26))/(1-(exp(-(-(v) -26)/9))) If v neq -26 | |
| hpower | 1.0 | |
| h Inf | 1 /(1+exp((v-(-65.0))/8.1)) | |
| h Tau | 0.40 + (0.265 * exp(-v/9.47)) | |
Nav1.3 is mainly expressed in the electrically excitable cells of the central nervous system (CNS) but is present at lower levels in other tissues and non-excitable cells [2117] [367].
Tissue and Cellular
Nav1.3 is widely expressed in the adult human central nervous system, in areas such as thalamus, amygdala, cerebellum, but is normally absent, or present at low levels, in the adult peripheral nervous system, such as the spinal chord and the heart. Via immunostaining of adult human brain tissue, Nav1.3 was found most present in the cerebral cortex with highest levels in the cerebellum. Nav1.3 was also identified in the gray matter of the middle frontal gyrus, middle temporal gyrus, and the sensory and motor cortex. Nav1.3 was also found present in the deep brain nuclei, the hippocampal structures and the insular cortex [1398].
Developmental
Across multiple species, the expression of scn3a is at its highest neonatally and post-birth, with scn3a mRNA levels dropping off over an organism’s lifetime [2113]. In the developing fetal brain, scn3a is present in the cortical plate, outer subventricular zone, intermediate zone and at lower expression levels in the ventricular zone [2101].
On a subcellular level, Nav1.3 was shown to be predominantly localized at the neuronal soma and proximal dendrites in the human brain [1398].
Cellular Function
Like all voltage gated sodium channels, Nav1.3 is responsible for the inward flow of Na+ ions into the neuronal cell in a voltage dependent manner. This depolarizing activity of Nav1.3 is likely involved in the establishment of the action potentials. Given their somatodendritic localization, it is thought that Nav1.3 ion channels participate in the downstream propagation of the action potential as that area controls neuronal excitability via the integration of synaptic impulses [2118]
Channelopathies
The other functions of Nav1.3 are highlighted by the different channelopathies that arise when the channel does not function as normal.
Neuropathic Pain
Nav1.3 is thought to play a role in neuropathic chronic pain. Though Nav1.3 expression is generally low in adults, Nav1.3 is re-expressed and up-regulated following spinal cord injury. Nav upregulation leads to the generation of ramp current, enhanced persistent current, and shifts in the activation and inactivation activity. These changes lead to increased firing of the downstream neurons leading to chronic pain in the injured area [2119].
Epilepsy
Numerous studies have identified mutations in scn3a that may be responsible for certain forms of epilepsy. Most mutations result in gain-of-function activity, where mutant channels inactivate slower and have a lower activation threshold. This results in increased misfiring rates. This effect can be hindered via application of sodium channel blockers [2120].
Development and associated disorders
Several studies have observed development disorders, such as cerebral cortex malformations, familial autism, oral motor development, linked to scn3a. Given that Nav1.3 is predominantly expressed in early development, mutations in scn3a are likely to lead to issues during this crucial stages. Indeed, altered scn3a expression disrupts the cerebral cortical development in ferret model experiments. [2101] [[1378].
Though it can function independently, the α subunit Nav1.3 often interacts with one or more β subunits through specific binding sites.
Navβ1 [Nav1.3] [Navβ1] complex expressed in xenopus oocytes was shown to increase peak current and shift in activation/inactivation activity, with a decrease in activation time and an increase in inactivation. This highlights the increased activation properties of the interaction and the potential of increased neuronal firing [44].
Navβ2 When [Nav1.3] [Navβ2] complex is expressed in mammalian systems, little to no effect was observed on the kinetics or current densities. However, when the same complex was expressed in DRG cells, a depolarizing shift in activation and faster recovery from inactivation was observed. This suggests that other interactions within the native cells are involved in the modulation of Nav1.3 activity in complex with Navβ2 [2114].
Navβ3 Navβ3 has been shown to be co-localised with Nav1.3 during the same stages of development in rats. Furthermore, Navβ3 was shown to interact with Nav1.3 resulting in slower inactivation rates and a hyperpolarization shift in the voltage dependence of activation and inactivation [46].
Tetrodotoxin (TTX) sensitivity is a key factor for differentiating ion channels. Nav1.3 is sensitive to tetrodotoxin, a toxin found in pufferfish. TTX works as a pore blocker to the ion channel by forming electrostatic interactions with the negatively charged vestibule of the Nav channel. This physically blocks the entrance to the pore, effectively stopping the flow of ions and current [2111].
Many studies have demonstrated the interactions between Nav1.3 and various compounds with the potential to modulate its activity. Certain of these compounds could be of pharmacological interest for further drug development.
For additional resources on potential drug and compound interactions:
References
Thimmapaya R
et al.
Distribution and functional characterization of human Nav1.3 splice variants.
Eur. J. Neurosci.,
2005
Jul
, 22 (1-9).
Tan J
et al.
Human and rat Nav1.3 voltage-gated sodium channels differ in inactivation properties and sensitivity to the pyrethroid insecticide tefluthrin.
Neurotoxicology,
2009
Jan
, 30 (81-9).
Cummins TR
et al.
Nav1.3 sodium channels: rapid repriming and slow closed-state inactivation display quantitative differences after expression in a mammalian cell line and in spinal sensory neurons.
J. Neurosci.,
2001
Aug
15
, 21 (5952-61).
Wang YW
et al.
Modulatory effect of auxiliary beta1 subunit on Nav1.3 voltage-gated sodium channel expressed in Xenopus oocyte.
Chin. Med. J.,
2007
Apr
20
, 120 (721-3).
Shah BS
et al.
Contactin associates with sodium channel Nav1.3 in native tissues and increases channel density at the cell surface.
J. Neurosci.,
2004
Aug
18
, 24 (7387-99).
Shah BS
et al.
Developmental expression of the novel voltage-gated sodium channel auxiliary subunit beta3, in rat CNS.
J. Physiol. (Lond.),
2001
Aug
1
, 534 (763-76).
Lindia JA
et al.
Distribution of the voltage gated sodium channel Na(v)1.3-like immunoreactivity in the adult rat central nervous system.
Brain Res.,
2003
Jan
17
, 960 (132-41).
Malo D
et al.
Three brain sodium channel alpha-subunit genes are clustered on the proximal segment of mouse chromosome 2.
Genomics,
1991
Jul
, 10 (666-72).
Meadows LS
et al.
Functional modulation of human brain Nav1.3 sodium channels, expressed in mammalian cells, by auxiliary beta 1, beta 2 and beta 3 subunits.
Neuroscience,
2002
, 114 (745-53).
Alessandri-Haber N
et al.
Molecular determinants of emerging excitability in rat embryonic motoneurons.
J. Physiol. (Lond.),
2002
May
15
, 541 (25-39).
Schaller KL
et al.
Expression and distribution of voltage-gated sodium channels in the cerebellum.
Cerebellum,
2003
, 2 (2-9).
Vega AV
et al.
L-type calcium channel activation up-regulates the mRNAs for two different sodium channel alpha subunits (Nav1.2 and Nav1.3) in rat pituitary GH3 cells.
Brain Res. Mol. Brain Res.,
2003
Aug
19
, 116 (115-25).
Jarnot M
et al.
Immunolocalization of NaV1.2 channel subtypes in rat and cat brain and spinal cord with high affinity antibodies.
Brain Res.,
2006
Aug
30
, 1107 (1-12).
Wood JN
et al.
Voltage-gated sodium channels and pain pathways.
J. Neurobiol.,
2004
Oct
, 61 (55-71).
Huang X
et al.
[Expression and function of voltage-gated Na+ channel isoforms in rat sinoatrial node]
Nan Fang Yi Ke Da Xue Xue Bao,
2007
Jan
, 27 (52-5).
Fry M
et al.
Differentiated pattern of sodium channel expression in dissociated Purkinje neurons maintained in long-term culture.
J. Neurochem.,
2007
May
, 101 (737-48).
Black JA
et al.
Sodium channel activity modulates multiple functions in microglia.
Glia,
2009
Aug
1
, 57 (1072-81).
Cusdin FS
et al.
The sodium channel {beta}3-subunit induces multiphasic gating in Nav1.3 and affects fast inactivation via distinct intracellular regions.
,
2010
Jul
30
, ().
Mechaly I
et al.
Molecular diversity of voltage-gated sodium channel alpha subunits expressed in neuronal and non-neuronal excitable cells.
Neuroscience,
2005
, 130 (389-96).
Weiss LA
et al.
Sodium channels SCN1A, SCN2A and SCN3A in familial autism.
Mol. Psychiatry,
2003
Feb
, 8 (186-94).
Waxman SG
et al.
Type III sodium channel mRNA is expressed in embryonic but not adult spinal sensory neurons, and is reexpressed following axotomy.
J. Neurophysiol.,
1994
Jul
, 72 (466-70).
Rogers M
et al.
The role of sodium channels in neuropathic pain.
Semin. Cell Dev. Biol.,
2006
Oct
, 17 (571-81).
Waxman SG
et al.
Anoxic injury of rat optic nerve: ultrastructural evidence for coupling between Na+ influx and Ca(2+)-mediated injury in myelinated CNS axons.
Brain Res.,
1994
May
2
, 644 (197-204).
Black JA
et al.
Upregulation of a silent sodium channel after peripheral, but not central, nerve injury in DRG neurons.
J. Neurophysiol.,
1999
Nov
, 82 (2776-85).
Whitaker WR
et al.
Comparative distribution of voltage-gated sodium channel proteins in human brain.
Brain Res. Mol. Brain Res.,
2001
Mar
31
, 88 (37-53).
Lindia JA
et al.
Relationship between sodium channel NaV1.3 expression and neuropathic pain behavior in rats.
Pain,
2005
Sep
, 117 (145-53).
Yu S
et al.
Upregulated expression of voltage-gated sodium channel Nav1.3 in cortical lesions of patients with focal cortical dysplasia type IIb.
Neuroreport,
2012
May
9
, 23 (407-11).
Holland KD
et al.
Mutation of sodium channel SCN3A in a patient with cryptogenic pediatric partial epilepsy.
Neurosci. Lett.,
2008
Mar
5
, 433 (65-70).
Nassar MA
et al.
Nerve injury induces robust allodynia and ectopic discharges in Nav1.3 null mutant mice.
,
2006
, 2 (33).
Smith RS
et al.
Sodium Channel SCN3A (NaV1.3) Regulation of Human Cerebral Cortical Folding and Oral Motor Development.
Neuron,
2018
Sep
05
, 99 (905-913.e7).
Jiang D
et al.
Structural Advances in Voltage-Gated Sodium Channels.
Front Pharmacol, 2022, 13 (908867).
Kasai N
et al.
Genomic structures of SCN2A and SCN3A - candidate genes for deafness at the DFNA16 locus.
Gene,
2001
Feb
7
, 264 (113-22).
Heighway J
et al.
Sodium channel expression and transcript variation in the developing brain of human, Rhesus monkey, and mouse.
Neurobiol Dis, 202203, 164 (105622).
Chahine M
et al.
Regulatory Role of Voltage-Gated Na Channel β Subunits in Sensory Neurons.
Front Pharmacol,
2011
, 2 (70).
Zhang J
et al.
N-type fast inactivation of a eukaryotic voltage-gated sodium channel.
Nat Commun, 20220517, 13 (2713).
Liao S
et al.
Structure and Function of Sodium Channel Nav1.3 in Neurological Disorders.
Cell Mol Neurobiol, 2022Mar24, ().
Black JA
et al.
Noncanonical roles of voltage-gated sodium channels.
Neuron,
2013
Oct
16
, 80 (280-91).
Wang J
et al.
Distribution and function of voltage-gated sodium channels in the nervous system.
Channels (Austin), 2017Nov02, 11 (534-554).
Hains BC
et al.
Sodium channel expression and the molecular pathophysiology of pain after SCI.
Prog. Brain Res.,
2007
, 161 (195-203).
Ademuwagun IA
et al.
Voltage Gated Sodium Channel Genes in Epilepsy: Mutations, Functional Studies, and Treatment Dimensions.
Front Neurol, 2021, 12 (600050).
Xu Q
et al.
Deglycosylation altered the gating properties of rNav1.3: glycosylation/deglycosylation homeostasis probably complicates the functional regulation of voltage-gated sodium channel.
,
2008
Oct
, 24 (283-7).
Rougier JS
et al.
Molecular determinants of voltage-gated sodium channel regulation by the Nedd4/Nedd4-like proteins.
Am. J. Physiol., Cell Physiol.,
2005
Mar
, 288 (C692-701).
Li X
et al.
Structural basis for modulation of human NaV1.3 by clinical drug and selective antagonist.
Nat Commun, 2022Mar11, 13 (1286).
Gustafson TA
et al.
Mutually exclusive exon splicing of type III brain sodium channel alpha subunit RNA generates developmentally regulated isoforms in rat brain.
J. Biol. Chem.,
1993
Sep
5
, 268 (18648-53).
Contributors:Katherine Johnston, Rajnish Ranjan, Michael Schartner
To cite this page: [Contributors] Channelpedia https://channelpedia.epfl.ch/wikipages/122/ , accessed on 2026 Mar 08