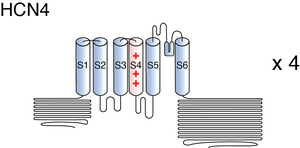
HCN4
Description: hyperpolarization activated cyclic nucleotide-gated potassium channel 4 Gene: Hcn4 Alias: HCN4, hcn4, Bcng3, BCNG-3, ih4
HCN4, encoded by the gene hcn4, is a hyperpolarization-activated cyclic nucleotide-gated potassium channel. HCN4 is predominantly expressed in cardiac pacemaking cells. It is involved in the generation of the I(f) current which controls cellular pacemaking activity.
Experimental data
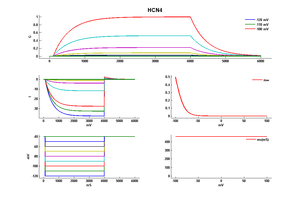
Rat HCN4 gene in CHO host cells |
||
|
Click for details 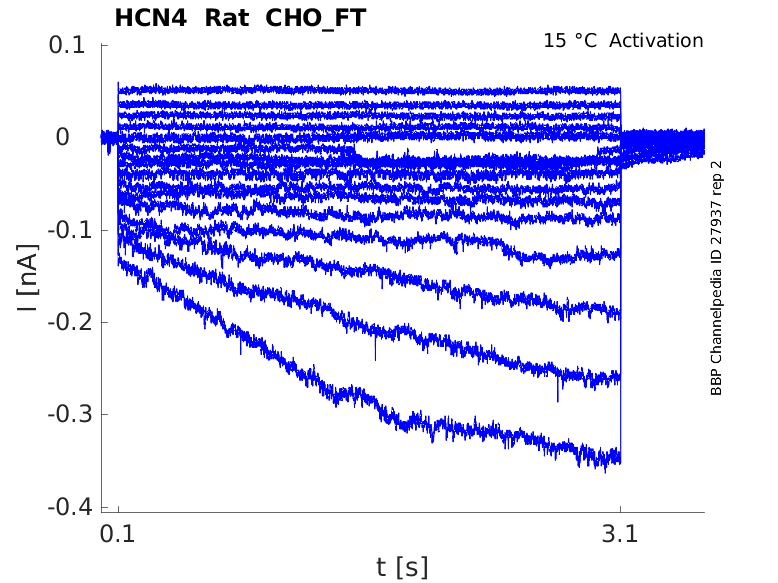
15 °Cshow 33 cells |
Click for details 
25 °Cshow 89 cells |
Click for details 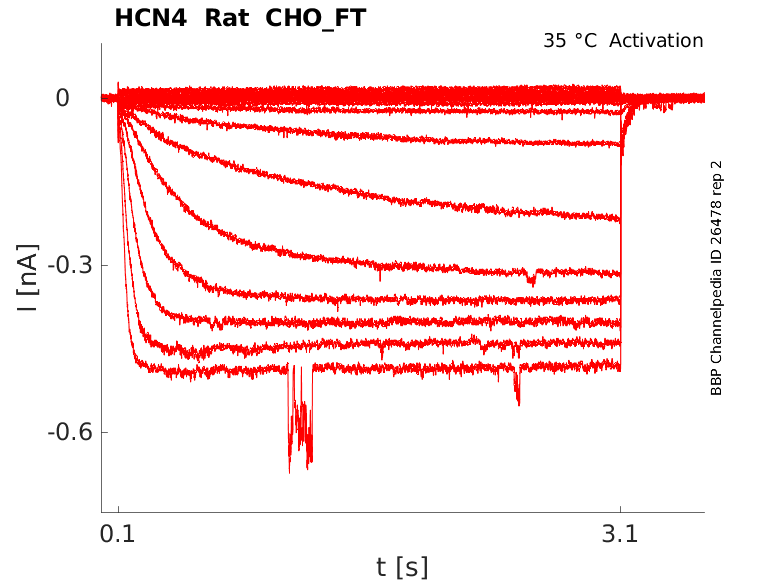
35 °Cshow 54 cells |
Mouse HCN4 gene in CHO host cells |
||
|
Click for details 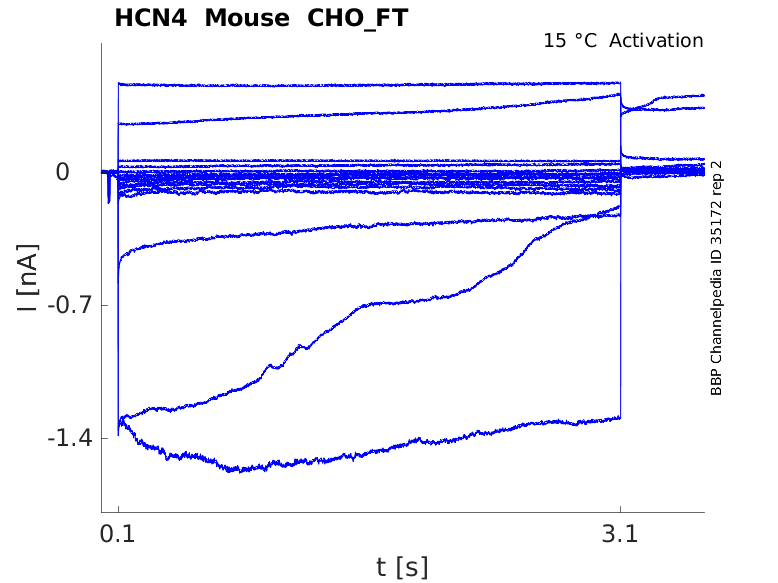
15 °Cshow 27 cells |
Click for details 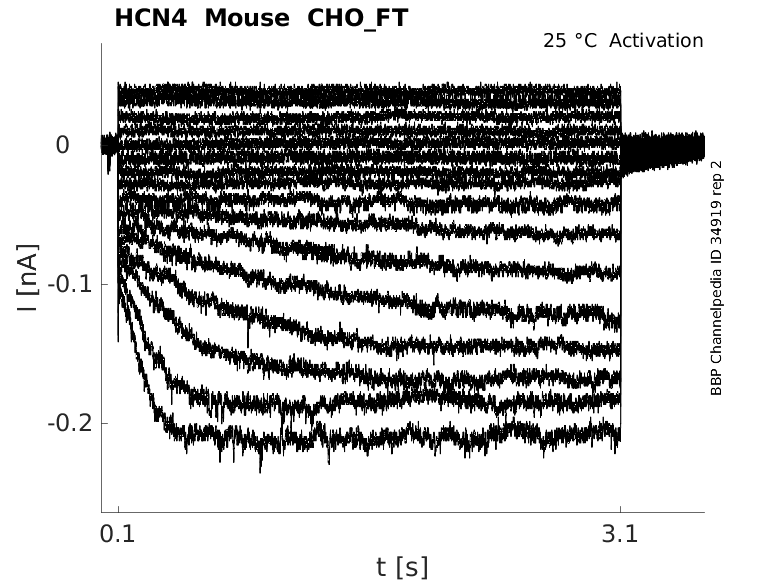
25 °Cshow 38 cells |
Click for details 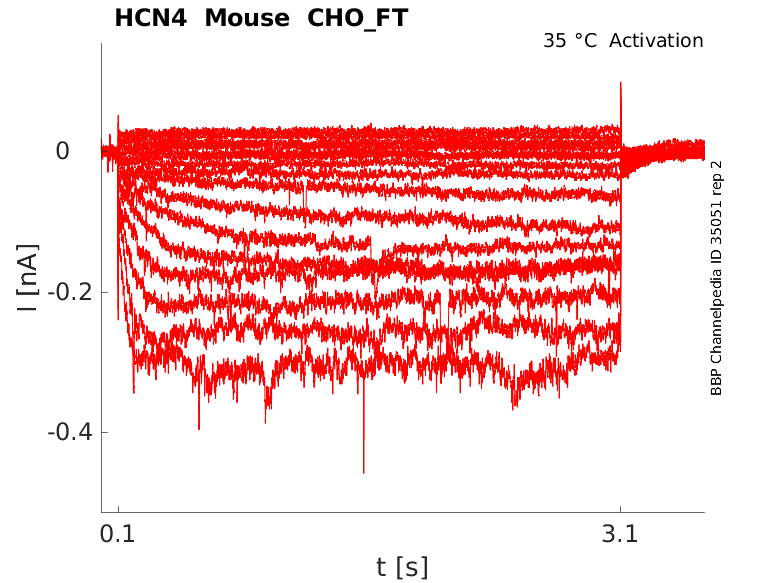
35 °Cshow 33 cells |
Human HCN4 gene in CHO host cells |
||
|
Click for details 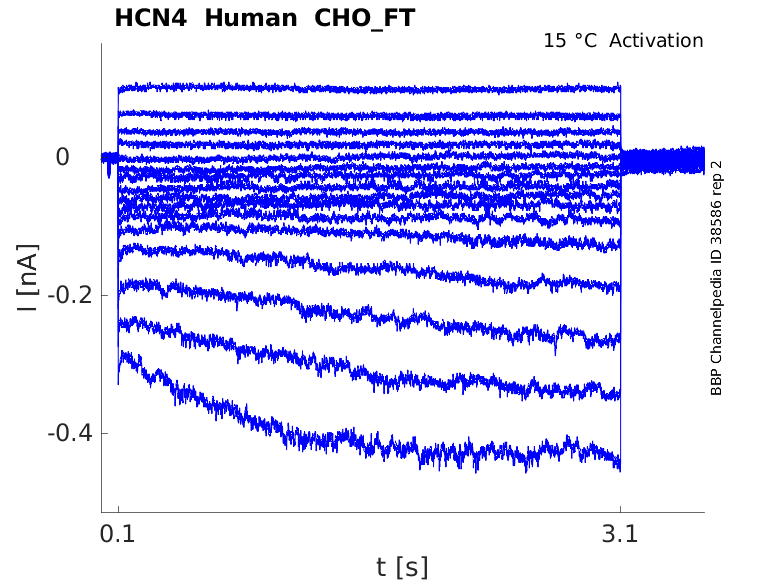
15 °Cshow 33 cells |
Click for details 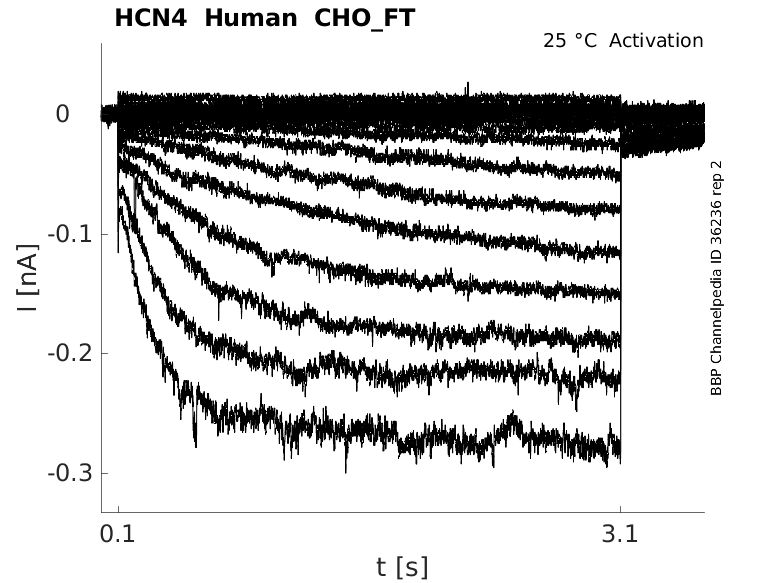
25 °Cshow 43 cells |
Click for details 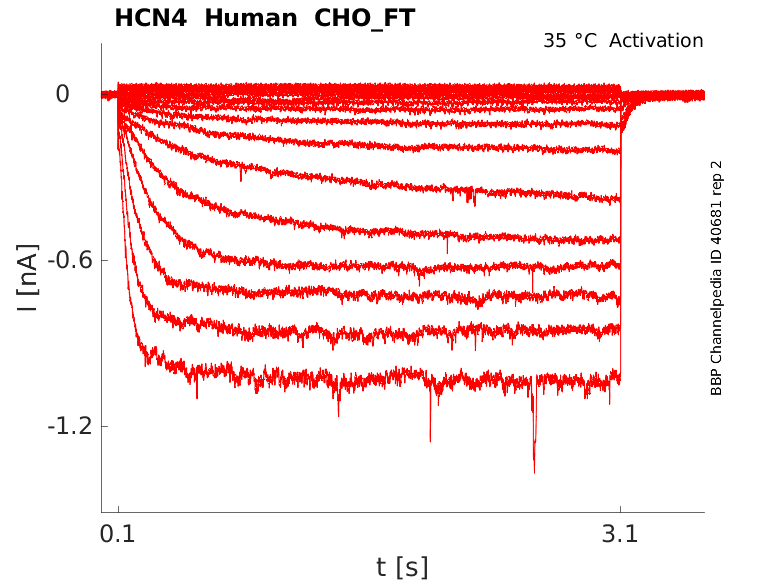
35 °Cshow 44 cells |
hcn4 is the coding gene for HCN4. In humans, hcn4 is located on chromosome 15q24.1 and is made up of 8 exons, all of which are coding.
To date, no transcript variants for hcn4 have been identified.
| Species | NCBI accession | Length (nt) | |
|---|---|---|---|
| Human | NM_005477.3 | 6922 | |
| Mouse | NM_001081192.3 | 6581 | |
| Rat | NM_021658.2 | 3947 |
The human HCN4 protein is composed of 1203 amino acid (aa) and has a molecular weight of 129 Kda.
Isoforms
HCN4 is subject to phosphorylation by serine/threonine kinases (p38 mitogen-activated protein kinase) and by tyrosine kinases (Src). The specific phosphorylation event involving the Y554 residue of hHCN4 by Src tyrosine kinase was shown to elicit an enhanced rate of channel activation and a concomitant shift in the current activation curve towards more positive membrane potentials. [2327] [457]
HCN4 is also subject to S-palmitoylation. Removal of S-palmitoylation sites does not exert a substantial influence on the kinetics of the channel but the PTM does appear to enhance the magnitude of HCN4-mediated currents [2298] [2345].
Visual Representation of HCN4 Structure
Methodology for visual representation of structure available here
The structure of rabbit HCN4 was resolved via cryo-electron microscopy to an overall resolution of 3.2 angstroms, giving us a detailed insight to the channel’s architecture. Structural resolution of the ion channel highlighted a number of HCN4 specific features: [2346]
- The voltage sensing domain of one subunit was packed against the pore of the same subunit (non-swapped), while the cytosolic domains were swapped.
- Notable structural differences in the S4-S5 linker and the C-linker.
- The possible sites for lipid interactions.
- Changes to the pore and selectivity filter that may encourage the passage of K+ over Na+ ions.
Other studies have highlighted the differences in the S1 and S1-S2 loop regions of HCN4 compared to HCN1, the consequences of which are discussed in the Kinetics section. [2347]. Divergences are also observed in various regions of the protein, including the S4-S5 linker, where the S4 segment undergoes a bending configuration after residue I405. This unique conformation enables interaction between the S3-S5 linker of one subunit and the B’ helix of the C-linker, leading to the formation of a tetrad structure, a feature not found in HCN1. [2346]
Additionally, HCN4 contains a unique pocket located between the C-linker and CNBD, identified as a likely binding site for di-cyclic nucleotides and the compound BPU. [2306]
HCN4 predicted AlphaFold size
Methodology for AlphaFold size prediction and disclaimer are available here
HCN4 has the slowest kinetics of the HCN channel family. Notably, the activation time constant of HCN4 is approximately 40-fold longer than that of HCN1, which represents the fastest among HCN channels when compared at a membrane potential of -100 mV. This discrepancy in kinetic speed is believed to arise from differences in the S1 transmembrane domain, the S1-S2 exoplasmic linker, and the S6-CNBD region. These specific regions are considered to be critical components influencing the gating activation of HCN channels. [457] [2348]
Similar to other HCN channels, HCN4 is believed to undergo mode shifting. However, the discernible effects of these mode shifts were primarily observed in the tail currents under specific conditions, such as in high-external K+ solutions. [2349] [457]
Single channel unitary conductance
There has been no single channel unitary conductance recordings for HCN4.
Model
Model HCN4 (ID=12)
| Animal | Mouse | |
| CellType | Dorsal root ganglion | |
| Age | 21 Days | |
| Temperature | 0.0°C | |
| Reversal | -45.0 mV | |
| Ion | Hcn + | |
| Ligand ion | ||
| Reference | [57] S Moosmang et. al; Eur. J. Biochem. 2001 Mar | |
| mpower | 1.0 | |
| m Inf | 1.0000/(1+exp((v- -100)/9.6)) | |
| m Tau | 461.0000 | |
HCN4 is found distributed across both the peripheral and central nervous system.
Extensive immunohistochemical studies focused on the localization of HCN channels have highlighted the presence of mRNA in the brain, including HCN4.[323] HCN4 channels are also expressed in select neurons in the brain, particularly neurons that show spontaneous rhythmic activity, such as thalamocortical relay neurons, dopaminergic neurons in the substantia nigra, cholinergic interneurons, and medial habenula neurons [2306]. Other CNS locations include olfactory bulb, purkinje fibers, distinct neurons of the basal ganglia, and mitral cell layer [338] [583] [2308] [2307] [457].
Within the PNS, HCN4 is expressed in the heart and is the major HCN isoform, accounting for ∼80% of I(h) in the heart [583]. It is particularly important in the sinoatrial node (SAN) and other mammalian cardiac tissues (32424620). In rabbits, mice, rats, and dogs, HCN4 is the main HCN isoform present in the SAN. In humans, HCN4 is the most highly expressed isoform in the SAN, atrioventricular node (AVN), atria, and ventricles (32424620).
For a more detailed map immunohistochemical localisation of HCN4, please consult the following paper [323]
The subcellular localization of HCN channels is neuron-type specific. It is therefore difficult to generalize and pinpoint the exact cellular locations of these channels.
In specific regions, such as the interneurons of the medial septum, hippocampus, and cerebellum, HCN4, in conjunction with other HCN channels, is expressed at both somatic and axonal regions. Additionally, HCN4 has been identified in receptor terminals of both myelinated and unmyelinated fibers in these areas. [2295] [2311]
HCN4 is the main pacemaker ion channel in the adult heart and contributes to the majority of the "funny current”, which is referred to as I(f) in the cardiac context. This contrasts with other tissues, where the "funny current" is typically denoted as I(h). [2350]
Pacemaking
Given its predominant expression in the heart, particularly in the pace sinoatrial node (SAN), HCN4 plays a crucial role in sustaining a steady cardiac rhythm. This is attributed to the significant contribution of I(f) to diastolic depolarization in pacemaker cells, which is fundamental for the initiation and regulation of the heart's rhythmic contractions. [2350]
As previously mentioned, HCN4 constitutes approximately 80% of the I(f) current in the sinoatrial (SA) node, which coincides with being the central node of cardiac pacemaker activity. Blocking HCN4 activity using ivabravine resulted in a complete loss of this current and was associated with rhythm disturbances [2327]. Furthermore, in cardiac-specific HCN4 knockout mice, it was observed that these mice did not survive during development due to heart arrest. This finding highlights the indispensable role of HCN4 in the proper generation of pacemaker potentials within the emerging sinoatrial node [1695] [2351]. In adult mice, the deletion of HCN4 resulted in cardiac arrhythmia, serving as further evidence that HCN4 is vital for maintaining a stable cardiac rhythm [2352].
HCN4 is believed to play a role in governing the day-night circadian rhythm in heart rate, although the precise mechanism underlying this phenomenon remains to be fully elucidated. [2353] [2354]
In the thalamus, the area of the brain where HCN4 is most present, HCN4 was shown to be essential for oscillatory activity in the thalamocortical (TC) network. HCN4-deficient TC neurons revealed a massive reduction of I(h) and strongly reduced intrinsic burst firing and significantly slowed down thalamic and cortical oscillations during active wakefulness.
Within the thalamus, a region where HCN4 is prominently expressed, HCN4 has been demonstrated to be essential for the generation of oscillatory activity in the thalamocortical (TC) network. In HCN4-deficient TC neurons, there was a substantial reduction in the I(h) current, resulting in a significant decrease in intrinsic burst firing. This deficiency led to a pronounced slowing of thalamic and cortical oscillations, particularly during active wakefulness. These findings accentuate the pivotal role of HCN4 in regulating the oscillatory dynamics within the thalamocortical network.[2355]
Channelopathies
Given the channel’s important role in the heart and pacemaking, mutations to HCN4 channel have been associated with several diseases and rhythm disturbances. Cardiac channelopathies associated with HCN4 are:
- Bradycardia [2327]
- Atrial Fibrillation [2327]
- Sinus Bradycardia [2356] [2357]
- Left Ventricular Noncompaction Cardiomyopathy (LVNC) [2356] [2357]
- Dilated Cardiomyopathy [2358]
Pathogenic variants of HCN4 have also been linked with epilepsy. Both gain-of-function (GOF) and loss-of-function (LOF) variants have been identified [2319].
HCN4, like other channels, is regulated by various auxiliary proteins and secondary messengers. These regulatory proteins play important roles in the development, localization, and expression of the channel protein.
Cyclic nucleotides
HCN4 and HCN2 both are highly sensitive to cyclic adenosine monophosphate (cAMP), which binds to the protein via the CNBD, located in the intracellular C-terminal. Binding of cAMP accelerates channel opening and shifts the activation curve to more positive potential. For HCN4, this varies between 10 and 25 mV. [2293] Other cyclic nucleotides also modulate the activity of HCN4:
- cCMP shifts the activation curve to more positive potentials, speeds up current activation, and decreases current deactivation. The voltage shift and the maximal increase in the current amplitude were significantly smaller than those observed for cAMP [2293].
- Pyrimidine cyclic nucleotide cCMP: cCMP shifts the activation curves of HCN2 and HCN4 to more depolarized voltages, speeds up activation, and slows down deactivation kinetics of these channels. [2302]
- pCPT-cGMP shifts hHCN4 activation to more positive potentials [56].
Other modulators
Shugoshin-1, a protein involved in chromosome segregation, interacts with HCN4 to promote and stabilize cardiac pacing, enhancing funny current and optimizing HCN4 cell-surface expression and function [2350].
Nkx2-5 suppresses the expression of HCN4 [2359].
HCN4 is sensitive to extracellular pH. Acidification to pH 3.9 was shown to speed up channel kinetics, right shifting the activation threshold to more positive potentials and inducing a depolarising shift of up to +35 mV. However, the mechanisms that cause this phenomenon are still unknown. [457]
HCN4 is sensitive to Cl−, as with HCN2. The regulation of HCN4 by Cl− thought to be relevant in heart (patho-)physiology [457].
MiRP1 has been shown to increase current densities of HCN4, but it also slows down kinetics and induces a negative shift of the activation curve of this channel [457].
HCN4 interacts with caveolin-3, a marker protein for caveolae. Clustering of HCN4 in caveolae has been proposed to be essential for its normal function and regulation [457].
For further compounds interactions, please consult the following resource
References
Brandt MC
et al.
Effects of KCNE2 on HCN isoforms: distinct modulation of membrane expression and single channel properties.
Am. J. Physiol. Heart Circ. Physiol.,
2009
Jul
, 297 (H355-63).
Stieber J
et al.
Functional expression of the human HCN3 channel.
J. Biol. Chem.,
2005
Oct
14
, 280 (34635-43).
Moosmang S
et al.
Cellular expression and functional characterization of four hyperpolarization-activated pacemaker channels in cardiac and neuronal tissues.
Eur. J. Biochem.,
2001
Mar
, 268 (1646-52).
Notomi T
et al.
Immunohistochemical localization of Ih channel subunits, HCN1-4, in the rat brain.
J. Comp. Neurol.,
2004
Apr
5
, 471 (241-76).
Moosmang S
et al.
Differential distribution of four hyperpolarization-activated cation channels in mouse brain.
Biol. Chem.,
1999 Jul-Aug
, 380 (975-80).
Biel M
et al.
Hyperpolarization-activated cation channels: from genes to function.
Physiol. Rev.,
2009
Jul
, 89 (847-85).
Gaborit N
et al.
Regional and tissue specific transcript signatures of ion channel genes in the non-diseased human heart.
J. Physiol. (Lond.),
2007
Jul
15
, 582 (675-93).
Benarroch EE
HCN channels: function and clinical implications.
Neurology,
2013
Jan
15
, 80 (304-10).
Baruscotti M
et al.
Deep bradycardia and heart block caused by inducible cardiac-specific knockout of the pacemaker channel gene Hcn4.
Proc. Natl. Acad. Sci. U.S.A.,
2011
Jan
25
, 108 (1705-10).
Sartiani L
et al.
The Hyperpolarization-Activated Cyclic Nucleotide-Gated Channels: from Biophysics to Pharmacology of a Unique Family of Ion Channels.
Pharmacol Rev, 2017Oct, 69 (354-395).
He C
et al.
Neurophysiology of HCN channels: From cellular functions to multiple regulations.
Prog. Neurobiol.,
2014
Jan
, 112 (1-23).
Itoh M
et al.
The hyperpolarization-activated cyclic nucleotide-gated (HCN) channels contain multiple S-palmitoylation sites.
J Physiol Sci, 2016May, 66 (241-8).
Zong X
et al.
Regulation of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel activity by cCMP.
J. Biol. Chem.,
2012
Aug
3
, 287 (26506-12).
Santoro B
et al.
Hyperpolarization-Activated Cyclic Nucleotide-Gated Channels as Drug Targets for Neurological Disorders.
Annu Rev Pharmacol Toxicol, 2020Jan06, 60 (109-131).
Shah MM
Cortical HCN channels: function, trafficking and plasticity.
J. Physiol. (Lond.),
2014
Jul
1
, 592 (2711-9).
Santoro B
et al.
Molecular and functional heterogeneity of hyperpolarization-activated pacemaker channels in the mouse CNS.
J. Neurosci.,
2000
Jul
15
, 20 (5264-75).
Doan TN
et al.
Differential distribution and function of hyperpolarization-activated channels in sensory neurons and mechanosensitive fibers.
J. Neurosci.,
2004
Mar
31
, 24 (3335-43).
Kessi M
et al.
The Contribution of HCN Channelopathies in Different Epileptic Syndromes, Mechanisms, Modulators, and Potential Treatment Targets: A Systematic Review.
Front Mol Neurosci, 2022, 15 (807202).
Congreve SD
et al.
Palmitoylation regulates the magnitude of HCN4-mediated currents in mammalian cells.
Front Physiol, 2023, 14 (1163339).
Saponaro A
et al.
Gating movements and ion permeation in HCN4 pacemaker channels.
Mol Cell, 2021Jul15, 81 (2929-2943.e6).
Ishii TM
et al.
Tryptophan-scanning mutagenesis in the S1 domain of mammalian HCN channel reveals residues critical for voltage-gated activation.
J. Physiol. (Lond.),
2007
Mar
1
, 579 (291-301).
Ishii TM
et al.
Determinants of activation kinetics in mammalian hyperpolarization-activated cation channels.
J. Physiol. (Lond.),
2001
Nov
15
, 537 (93-100).
Elinder F
et al.
Mode shifts in the voltage gating of the mouse and human HCN2 and HCN4 channels.
J. Physiol. (Lond.),
2006
Sep
1
, 575 (417-31).
Liu D
et al.
Cohesin-protein Shugoshin-1 controls cardiac automaticity via HCN4 pacemaker channel.
Nat Commun, 2021May05, 12 (2551).
Stieber J
et al.
The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart.
Proc. Natl. Acad. Sci. U.S.A.,
2003
Dec
9
, 100 (15235-40).
Herrmann S
et al.
HCN4 provides a 'depolarization reserve' and is not required for heart rate acceleration in mice.
EMBO J.,
2007
Oct
31
, 26 (4423-32).
Anderson C
et al.
Non-canonical role of the sympathetic nervous system in the day-night rhythm in heart rate.
Philos Trans R Soc Lond B Biol Sci, 2023Jun19, 378 (20220179).
D'Souza A
et al.
A circadian clock in the sinus node mediates day-night rhythms in Hcn4 and heart rate.
Heart Rhythm, 2021May, 18 (801-810).
Zobeiri M
et al.
The Hyperpolarization-Activated HCN4 Channel is Important for Proper Maintenance of Oscillatory Activity in the Thalamocortical System.
Cereb Cortex, 2019May01, 29 (2291-2304).
Milano A
et al.
HCN4 mutations in multiple families with bradycardia and left ventricular noncompaction cardiomyopathy.
J. Am. Coll. Cardiol.,
2014
Aug
26
, 64 (745-56).
DiFrancesco D
Funny channel gene mutations associated with arrhythmias.
J. Physiol. (Lond.),
2013
Sep
1
, 591 (4117-24).
Yampolsky P
et al.
Augmentation of myocardial If dysregulates calcium homeostasis and causes adverse cardiac remodeling.
Nat Commun, 2019Jul23, 10 (3295).
Mommersteeg MT
et al.
Molecular pathway for the localized formation of the sinoatrial node.
Circ. Res.,
2007
Feb
16
, 100 (354-62).
Contributors: Katherine Johnston, Rajnish Ranjan, Michael Schartner, Nitin Khanna
To cite this page: [Contributors] Channelpedia https://channelpedia.epfl.ch/wikipages/64/ , accessed on 2026 Feb 25