Nav
Description: Sodium channel
Voltage-gated sodium channels are responsible for action potential initiation and propagation in excitable cells, including nerve, muscle, and neuroendocrine cell types. They are also expressed at low levels in nonexcitable cells, where their physiological role is unclear. Sodium channels are the founding members of the ion channel superfamily in terms of their discovery as a protein and determination of their amino acid sequence. [817]
Four sodium channel isoforms (Nav1.1, Nav1.2, Nav1.3 and Nav1.6) are highly expressed in the central nervous system. Two isoforms are abundant in muscle: Nav1.4 in adult skeletal muscle and Nav1.5 in embryonic and denervated skeletal muscle and heart muscle. Finally, Nav1.7, Nav1.8 and Nav1.9, are expressed primarily in the peripheral nervous system [834].
In mammals, nine functionally expressed voltage-gated sodium channel (Nav1.1-Nav1.9) are encoded by 9 genes [834]. These are, respectively:
- Nav1.1 : scn1a
- Nav1.2 : scn2a
- Nav1.3 : scn3a
- Nav1.4 : scn4a
- Nav1.5 : scn5a
- Nav1.6 : scn8a
- Nav1.7 : scn9a
- Nav1.8 : scn10a
- Nav1.9 : scn11a
Sodium channels were the first members of the ion channel superfamily to be discovered; the superfamily also includes voltage-gated K+ channels, voltage-gated Ca2+ channels, Trp-related channels (a diverse family permeable to various cations) and cyclic-nucleotide-gated channels. However, in evolution, the sodium channel family is the most recent of the voltage-gated ion channels to have arisen, having evolved from similarly structured Ca2+ channels that contain four homologous domains.
For a phylogenetic tree and nomenclature/classification, see Catterall [817].
All voltage gated sodium have a canonical mRNA transcript coding for the canonical ion channel protein.
However, depending on the VGSC subtype, there exist a number of transcript variants as a result of alternative splicing. Alternative splicing allows for greater diversity of protein function and neuronal responses, thus expanding neuronal function and network structure. source
Though some splicing events are specific to certain Navs, one event is conserved across most Nav subtypes: The alternative splicing of exon 5 neonatal (N) and adult (A). These transcript variants are generated via the inclusion of either exon 5N or exon 5A, respectively.
When this splicing event was first identified, the transcripts found during development were coined as “neonatal” in contrast to the “adult” transcripts. Though initially named based on these developmental expression observations, the presence of exon 5A or exon 5N in a transcript depends on the specific ion channel and is not always developmentally regulated. For human scn2a, scn3a, scn8a, it is indeed developmentally regulated, with the expression of 5N or 5A being mutually exclusive. For scn1a, scn5a, scn9a, alternative 5N and 5A splicing occurs but it is not under developmental control. [2113]
When refers a particular Nav, one generally means the canonical isoforms.
However, depending on the Nav subtype, there exist a number of protein isoforms depending on the translation of transcript variants.
The isoform from the translation of the alternative splicing of exon 5 resulting in a protein with substitution in the loop between S3 and S4 of Domain I. source
Like all mammalian proteins, Navs are subject to post translational modification.
These include, but are not limited to:
- Phosphorylation
- Glycosylation
- Ubiquitination
- Arginine methylation
- Palmitoylation
- SUMOylation
- Nitrosylation
- Reactive oxygen species (ROS) modification
The functional effects of channel phosphorylation on NaV electrophysiology often depend on the specific Nav subtype. A lot of research on PTMs has highlighted how they can alter trafficking, localization, gating, and pharmacology, among other properties. [2233] [2145]
Visual Representation of Nav Structure
Methodology for visual representation of structure available here
For further structural overview images see Yu [820] and Catterall [1367].
The eukaryotic voltage gated sodium channel (Nav) is comprised of a large pore-forming α subunit, which mediates voltage dependent ion conductance, and one or two auxiliary β subunits that modulate channel activity [2107] [2108].
Overall, Nav channel subtypes are structurally conserved with some sequences variations between the different subtypes, conferring each ion channel it’s specific structure, expression pattern, and activity [2110] [2111].
The ion channel protein is also known as the α-subunit. This α-subunit has four repeat domains, labeled I through IV (DI-DIV), each containing six membrane-spanning regions, labelled S1 through S6.
The highly conserved S4 region acts as the channel's voltage sensor domain (VSD). The voltage sensitivity of this channel is due to positive amino acids located at every third position. When stimulated by a change in transmembrane voltage, this region moves toward the extracellular side of the cell membrane, allowing the channel to become permeable to ions.
The ions are conducted through a pore domain (PM), which can be broken into two regions. The more external (i.e., more extracellular) portion of the pore is formed by the "P-loops" (the region between S5 and S6) of the four domains. This region is the most narrow part of the pore and is responsible for its ion selectivity. Being only 0.3 by 0.5 nm wide, this area of the pore is just large enough to allow a single Na+ ion with a water molecule associated to pass through. This "selectivity filter" is made of negatively charged amino acid residues, which attract the positive Na+ ion and keep out negatively charged ions, such as chloride.The inner portion (i.e., more cytoplasmic) of the pore is formed by the combined S5 and S6 regions of the four domains.
The region linking domains III and IV, the IFM motif, is also important for channel function. The IFM motif binds to a hydrophobic receptor site next to the S6 in D4. This binding causes the shift of S6, allosterically closing the channel, thus deactivating the channel. [834]
Voltage gated sodium channels are known to have fast kinetics.
These channels activate rapidly in response to depolarization and conduct a large inward Na+ current, driving the rapid upstroke of the action potential [2147]. Fast inactivation quickly kicks in following the channel opening, with the protein returning to its resting state after repolarization. Most voltage gated sodium channels, upon prolonged depolarization, progressively enter the slow inactivation (SI) state from which they are also slow to recover. [820]
Given the specific properties each voltage gated sodium channel, individual Nav channels may also display other types of kinetics activity. For example, Nav1.1, Nav1.2, and Nav1.6 are know to display persistent current. Upon prolonged depolarisation, these channels remain open or partially open, allowing a sustained inward flow of sodium ions and thus extended duration of action potentials, enhanced neuronal firing. [834]
Single Channel Unitary Conductance
Single channel unitary conductance refers to the conductance of an individual ion channel when it is open and is typically measured in siemens (S) or pico siemens (pS). The conductance value is influenced by various factors, including the size and charge of the ions, as well as the properties of the channel itself, such as its structure, selectivity, and gating mechanisms.
The conductance can be determined through experimental techniques like patch-clamp electrophysiology, where a fine glass micropipette is used to isolate and measure the electrical currents flowing through individual ion channels.
Model
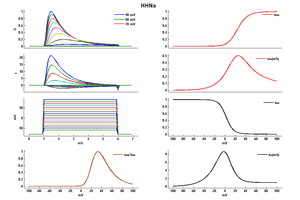
Model HHNa (ID=2)
| Animal | Squid | |
| CellType | giant Axon | |
| Age | 0 Days | |
| Temperature | 9.3°C | |
| Reversal | 50.0 mV | |
| Ion | Na + | |
| Ligand ion | ||
| Reference | [262] A L Hodgkin et. al; Bull. Math. Biol. 1990 | |
| mpower | 3.0 | |
| m Alpha | (0.1*(25-v))/(exp((25-v)/10) -1.0) If v neq 25 | |
| m Beta | 4.0 * (exp(-v/18)) | |
| hpower | 1.0 | |
| h Alpha | 0.07 * exp(-v/20) | |
| h Beta | 1/(exp((30-v)/10) + 1.0) | |
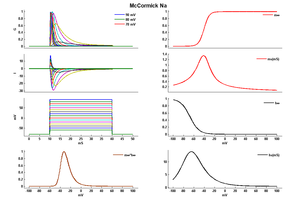
Model McCormick Na (ID=34)
| Animal | rat | |
| CellType | neocortical pyramidal | |
| Age | 14 Days | |
| Temperature | 23.0°C | |
| Reversal | 50.0 mV | |
| Ion | Na + | |
| Ligand ion | ||
| Reference | [283] D A McCormick et. al; J. Neurophysiol. 1992 Oct | |
| mpower | 3.0 | |
| m Alpha | 0.091*(v+38)/(1-exp((-v-38)/5)) If v neq -38 | |
| m Beta | -0.062*(v+38)/(1-exp((v+38)/5)) If v neq -38 | |
| hpower | 1.0 | |
| h Alpha | 0.016*exp((-55-v)/15) | |
| h Beta | 2.07/(exp((17-v)/21)+1) | |
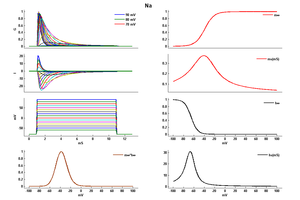
Model Na (ID=35)
| Animal | rat | |
| CellType | Neocortical | |
| Age | 0 Days | |
| Temperature | 23.0°C | |
| Reversal | 50.0 mV | |
| Ion | Na + | |
| Ligand ion | ||
| Reference | [284] J R Huguenard et. al; J. Neurophysiol. 1988 Mar | |
| mpower | 3.0 | |
| m Alpha | (0.182 * (v- -35))/(1-(exp(-(v- -35)/9))) If v neq -35 | |
| m Beta | (0.124 * (-v -35))/(1-(exp(-(-v -35)/9))) If v neq -35 | |
| hpower | 1.0 | |
| h Inf | 1.0/(1+exp((v- -65)/6.2)) | |
| h Tau | 1/((0.024 * (v- -50))/(1-(exp(-(v- -50)/5))) +(0.0091 * (-v - 75.000123))/(1-(exp(-(-v - 75.000123)/5)))) If v neq -50 | |
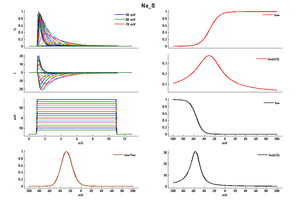
Model Na_S (ID=36)
| Animal | rat | |
| CellType | Neocortical | |
| Age | 0 Days | |
| Temperature | 23.0°C | |
| Reversal | 50.0 mV | |
| Ion | Na + | |
| Ligand ion | ||
| Reference | [285] J R Huguenard et. al; Cereb. Cortex 1991 Jan-Feb | |
| mpower | 3.0 | |
| m Alpha | (0.182 * ((v-10)- -35))/(1-(exp(-((v-10)- -35)/9))) If v neq -25 | |
| m Beta | (0.124 * (-(v-10) -35))/(1-(exp(-(-(v-10) -35)/9))) If v neq -25 | |
| hpower | 1.0 | |
| h Inf | 1.0/(1+exp((v- -65-10)/6.2)) | |
| h Tau | 1/((0.024 * ((v-10)- -50))/(1-(exp(-((v-10)- -50)/5))) +(0.0091 * (-(v-10) - 75.000123))/(1-(exp(-(-(v-10) - 75.000123)/5)))) If v neq -40 | |
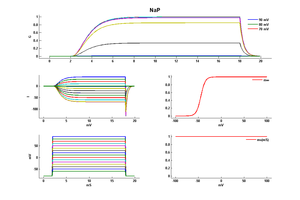
Model NaP (ID=37)
Persistent Sodium channel
| Animal | rat | |
| CellType | L5PC | |
| Age | 21 Days | |
| Temperature | 23.0°C | |
| Reversal | 50.0 mV | |
| Ion | Na + | |
| Ligand ion | ||
| Reference | [286] Nadav Astman et. al; J. Neurosci. 2006 Mar 29 | |
| mpower | 3.0 | |
| m Inf | 1.0000/(1+ exp((v - -44.0000)/-4.8500)) | |
| m Tau | 1.0 | |
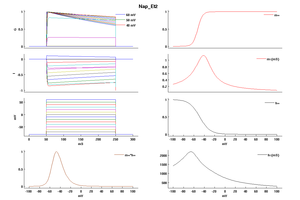
Model Nap_Et2 (ID=51)
| Animal | rat | |
| CellType | Neocortical L5PC | |
| Age | 0 Days | |
| Temperature | 23.0°C | |
| Reversal | 50.0 mV | |
| Ion | Na + | |
| Ligand ion | ||
| Reference | [1500] Costa M Colbert et. al; Nat. Neurosci. 2002 Jun | |
| mpower | 3.0 | |
| m Alpha | 1.0/(1+exp((v- -52.6)/-4.6)) | |
| m Beta | 6*(1/(((0.182 * (v- -38))/(1-(exp(-(v- -38)/6)))) + ((0.124 * (-v -38))/(1-(exp(-(-v -38)/6))))))/qt If v neq -38 | |
| hpower | 1.0 | |
| h Inf | 1.0/(1+exp((v- -48.8)/10)) | |
| h Tau | (1/((-2.88e-6 * (v + 17) / (1 - exp((v + 17)/4.63))) + (6.94e-6 * (v + 64.4) / (1 - exp(-(v + 64.4)/2.63)))))/qt If v neq -64.4 | |
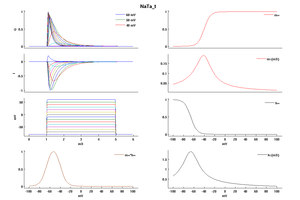
Model NaTa_t (ID=52)
| Animal | rat | |
| CellType | Neocortical L5PC | |
| Age | 0 Days | |
| Temperature | 23.0°C | |
| Reversal | 50.0 mV | |
| Ion | Na + | |
| Ligand ion | ||
| Reference | [1500] Costa M Colbert et. al; Nat. Neurosci. 2002 Jun | |
| mpower | 3.0 | |
| m Alpha | (0.182 * (v- -38))/(1-(exp(-(v- -38)/6))) If v neq -38 | |
| m Beta | (0.124 * (-v -38))/(1-(exp(-(-v -38)/6))) If v neq -38 | |
| hpower | 1.0 | |
| h Alpha | (-0.015 * (v- -66))/(1-(exp((v- -66)/6))) If v neq -66 | |
| h Beta | (-0.015 * (-v -66))/(1-(exp((-v -66)/6))) If v neq -66 | |
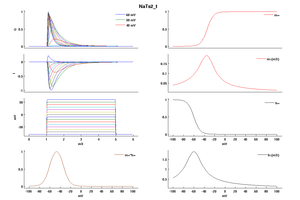
Model NaTs2_t (ID=53)
| Animal | rat | |
| CellType | Neocortical L5PC | |
| Age | 0 Days | |
| Temperature | 23.0°C | |
| Reversal | 50.0 mV | |
| Ion | Na + | |
| Ligand ion | ||
| Reference | [1500] Costa M Colbert et. al; Nat. Neurosci. 2002 Jun | |
| mpower | 3.0 | |
| m Alpha | (0.182 * (v- -32))/(1-(exp(-(v- -32)/6))) If v neq -32 | |
| m Beta | (0.124 * (-v -32))/(1-(exp(-(-v -32)/6))) If v neq -32 | |
| hpower | 1.0 | |
| h Alpha | (-0.015 * (v- -60))/(1-(exp((v- -60)/6))) If v neq -60 | |
| h Beta | (-0.015 * (-v -60))/(1-(exp((-v -60)/6))) If v neq -60 | |
Voltage gated sodium channels are primarily located in the nervous system. [834]
- Nav1.1, Nav1.2, Nav1.3, and Nav1.6 are expressed abundantly in the CNS
- Nav1.7, Nav1.8, and Nav1.9 are expressed primarily in the PNS
- Nav1.4 and Nav1.5 are found in contractile fibres of skeletal muscles and the cardiomyocytes respectively
Though predominantly found in those areas, the expression of the specific Nav is not restricted to that one location. Indeed, the distribution of an particular Nav will depend on its subtype. Please refer to the specific page for more detailed information.
The subcellular distribution of Navs will depend on the particular Nav subtype. Please refer to the specific page for more detailed information
Given their kinetics, voltage-gated sodium channels are mostly responsible for action potential initiation and propagation in excitable cells, including nerve, muscle, and neuroendocrine cell types. Therefore they contribute to the electrical signalling and excitability of these cells and tissues in which they are present.
Channelopathies [1385]
Mutations in any of the genes encoding the α-subunits can alter its biophysical properties of the channel, leading to the development of “channelopathies”. Channelopathies result from autosomal dominant inheritance and de novo mutations and can be classified in four groups depending on the predominant organ involved :
- Brain sodium channelopathies: these include mutations in scn1a, scn2a, scn3a and some mutations in scn8a. These mutations may give rise to epilepsy and epileptic/convulsive disorders.
- Skeletal muscle sodium channelopathies: involve mutations in the scn4a that are associated with myotonia, myasthenia syndromes, and paralysis.
- Cardiac sodium channelopathies: which involve mutations in scn5a and scn10a. They have been associated with alterations in the ventricular conduction.
- Peripheral nerve sodium channelopathies: include mutations in scn9a, scn10a, scn11a. Mutations in these genes have been associated with peripheral pain syndromes, including neuropathic and inflammatory pain.
Voltage-gated sodium channels are regulated by various auxiliary proteins and secondary messengers. These regulatory proteins play important roles in the development, localization, and expression of the channel protein.
β-subunit [834]
Some of the most important auxilary proteins are the beta subunits. To date, 5 β subunits have been identified, Navβ1, Navβ2, Navβ3, Navβ4, and Navβ1B a truncated form of β1 [878] [2109]. Though Navs can be expressed and function alone in vitro, interaction with a β subunit is crucial for the normal physiological activity of the protein.
All Navβ subunits contain immunoglobulin-like folds similar to those found in neural cell adhesion molecules. The sequences of the Navβ subunits are not homologous, but they are all clearly related based on the phylogenetic tree. Each of the beta subunit sequences predicts a protein with an amino-terminal signal sequence and single membrane-spanning region, indicative of an extracellular amino terminus.
The Navβ1 and Navβ3 subunits bing non-covalently to the α-subunit.
The Navβ2 and Navβ4 subunits is covalently link to the α-subunit by disulfide bonds.
Co-expression of the Navβ subunits along with an α-subunit leads to modulation of the latter protein :
- Navβ1 generally modulates the electrophysiological properties of the channel, including accelerating inactivation and shifting the voltage-dependence of steady-state inactivation in the negative direction.
- Navβ3 subunit modulates gating of the α-subunit sodium channels to a lesser extent than does co-expression of Navβ1
- Navβ2 subunit modulates alfa-subunit gating the least of the beta-subunits.
However, the interaction between the α- and Navβ subunits depends on the individual ion channel interaction. Please refer to the individual ion channel pages for more specific interact details.
The beta-subunits are also important for sodium channel interactions with cellular proteins. The Navβ2 subunit significantly increases membrane capacitance, which may indicate that it is involved in insertion of the channels into the cellular membrane. Both Navβ1 and Navβ2 interact with the extracellular matrix proteins tenascin-C and tenascin-R, suggesting that the proteins may function as cellular adhesion molecules. Consistent with this hypothesis, both Navβ1 and Navβ2 subunits recruit ankyrin to sites of cell-cell contact, and this recruitment requires the cytoplasmic domains of the subunits.
Toxins and other compounds
All of the pharmacological agents that act on sodium channels have receptor sites on the α subunits. At least six distinct receptor sites for neurotoxins and one receptor site for local anesthetics and related drugs have been identified [815] [817].
Neurotoxin receptor site 1 binds the nonpeptide pore blockers tetrodotoxin, saxitoxin, and the peptide pore blocker mu-conotoxin [818], [819], [815].
The different VGSC subtypes can also be easily distinguished on the basis of toxin sensitivity [834] :
- Sodium channels present in adult skeletal muscle are sensitive to nanomolar concentrations of both TTX and mu-conotoxin GIIIA.
- Sodium channels in CNS are sensitive to nanomolar concentration of TTX.
- Sodium channels expressed in cardiac muscle cells are resistant to nanomolar concentrations of TTX, and require micromolar concentrations for inhibition. On the other hand, these channels are more sensitive to inhibition by lidocaine than are CNS channels.
The receptor sites for these toxins are formed by amino acid residues in the pore loops and immediately on the extracellular side of the pore loops at the outer end of the pore:
Local anesthetics and related antiepileptic and antiarrhythmic drugs bind to overlapping receptor sites located in the inner cavity of the pore of the sodium channel [815]. Amino acid residues in the S6 segments from at least three of the four domains contribute to this complex drug receptor site, with the IVS6 segment playing the dominant role. Tables 2 through 10 in [817] summarize the major molecular, physiological, and pharmacological properties for each of the nine sodium channels that have been functionally expressed. Quantitative data are included for voltage dependence of activation and inactivation, single-channel conductance, and binding of drugs and neurotoxins, focusing on those agents that are widely used and diagnostic of channel identity and function.
The current density and gating properties of VGSC can also be modulated by the differential expression of channel “partners” or ChiPs. These include :
- Caveolin-3 (and the membrane compartment “caveolae”)
- CaMKII
- Connexin-43
- Telethonin
- Plakophilin
- Ankyrins
- Fibroblast growth factor homologous factors (FHFs)
- Nedd4
- SAPs
- Syntrophin/dystrophin complex.
For further information see Table 3 (VGSC protein partners) in [1376]
References
Hodgkin AL
et al.
A quantitative description of membrane current and its application to conduction and excitation in nerve. 1952.
Bull. Math. Biol.,
1990
, 52 (25-71; discussion 5-23).
Traub RD
et al.
Fast rhythmic bursting can be induced in layer 2/3 cortical neurons by enhancing persistent Na+ conductance or by blocking BK channels.
J. Neurophysiol.,
2003
Feb
, 89 (909-21).
McCormick DA
et al.
A model of the electrophysiological properties of thalamocortical relay neurons.
J. Neurophysiol.,
1992
Oct
, 68 (1384-400).
Huguenard JR
et al.
Developmental changes in Na+ conductances in rat neocortical neurons: appearance of a slowly inactivating component.
J. Neurophysiol.,
1988
Mar
, 59 (778-95).
Hamill OP
et al.
Patch-clamp studies of voltage-gated currents in identified neurons of the rat cerebral cortex.
Cereb. Cortex,
1991 Jan-Feb
, 1 (48-61).
Astman N
et al.
Persistent sodium current in layer 5 neocortical neurons is primarily generated in the proximal axon.
J. Neurosci.,
2006
Mar
29
, 26 (3465-73).
Kole MH
et al.
Action potential generation requires a high sodium channel density in the axon initial segment.
Nat. Neurosci.,
2008
Feb
, 11 (178-86).
Nusser Z
Variability in the subcellular distribution of ion channels increases neuronal diversity.
Trends Neurosci.,
2009
May
, 32 (267-74).
Catterall WA
From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels.
Neuron,
2000
Apr
, 26 (13-25).
Catterall WA
et al.
International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels.
Pharmacol. Rev.,
2005
Dec
, 57 (397-409).
Fozzard HA
et al.
Structure and function of voltage-dependent sodium channels: comparison of brain II and cardiac isoforms.
Physiol. Rev.,
1996
Jul
, 76 (887-926).
Terlau H
et al.
Structure and function of voltage-gated ion channels.
Naturwissenschaften,
1998
Sep
, 85 (437-44).
Shapiro L
et al.
Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin.
Neuron,
1996
Sep
, 17 (435-49).
Akopian AN
et al.
Structure and distribution of a broadly expressed atypical sodium channel.
FEBS Lett.,
1997
Jan
3
, 400 (183-7).
Zakon HH
Adaptive evolution of voltage-gated sodium channels: the first 800 million years.
Proc. Natl. Acad. Sci. U.S.A.,
2012
Jun
26
, 109 Suppl 1 (10619-25).
Catterall WA
Voltage-gated sodium channels at 60: structure, function and pathophysiology.
J. Physiol. (Lond.),
2012
Jun
1
, 590 (2577-89).
Savio-Galimberti E
et al.
Voltage-gated sodium channels: biophysics, pharmacology, and related channelopathies.
Front Pharmacol,
2012
, 3 (124).
George AL
Inherited disorders of voltage-gated sodium channels.
J. Clin. Invest.,
2005
Aug
, 115 (1990-9).
Hu F
et al.
17β-Estradiol regulates the gene expression of voltage-gated sodium channels: role of estrogen receptor α and estrogen receptor β.
Endocrine,
2012
Apr
, 41 (274-80).
Colbert CM
et al.
Ion channel properties underlying axonal action potential initiation in pyramidal neurons.
Nat. Neurosci.,
2002
Jun
, 5 (533-8).
Magistretti J
et al.
Biophysical properties and slow voltage-dependent inactivation of a sustained sodium current in entorhinal cortex layer-II principal neurons: a whole-cell and single-channel study.
J. Gen. Physiol.,
1999
Oct
, 114 (491-509).
Catterall WA
The molecular basis of neuronal excitability.
Science,
1984
Feb
17
, 223 (653-61).
O'Malley HA
et al.
Sodium channel β subunits: emerging targets in channelopathies.
Annu. Rev. Physiol.,
2015
, 77 (481-504).
Brackenbury WJ
et al.
Na Channel β Subunits: Overachievers of the Ion Channel Family.
Front Pharmacol,
2011
, 2 (53).
Noreng S
et al.
Structural Pharmacology of Voltage-Gated Sodium Channels.
J Mol Biol, 20210820, 433 (166967).
Jiang D
et al.
Structural Advances in Voltage-Gated Sodium Channels.
Front Pharmacol, 2022, 13 (908867).
Heighway J
et al.
Sodium channel expression and transcript variation in the developing brain of human, Rhesus monkey, and mouse.
Neurobiol Dis, 202203, 164 (105622).
Pei Z
et al.
Posttranslational Modification of Sodium Channels.
Handb Exp Pharmacol, 2018, 246 (101-124).
Onwuli DO
et al.
An update on transcriptional and post-translational regulation of brain voltage-gated sodium channels.
Amino Acids, 2016Mar, 48 (641-651).
Contributors: Rajnish Ranjan, Michael Schartner, Erika Borcel
To cite this page: [Contributors] Channelpedia https://channelpedia.epfl.ch/wikipages/189/ , accessed on 2026 Feb 22