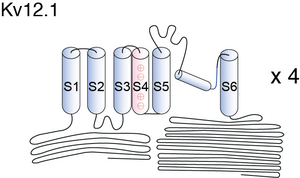
Kv12.1
Description: potassium voltage-gated channel, subfamily H (eag-related), member 8 Gene: Kcnh8 Alias: Kv12.1, ELK, ELK1, elk3, KCNH8
Kv12.1, encoded by the gene KCNH8, is a voltage-gated potassium channel, pore-forming (alpha) subunit, subfamily H. Kv12.1 is also known as ELK, ELK1, and elk3.
Experimental data
Rat Kv12.1 gene in CHO host cells datasheet |
||
|
Click for details 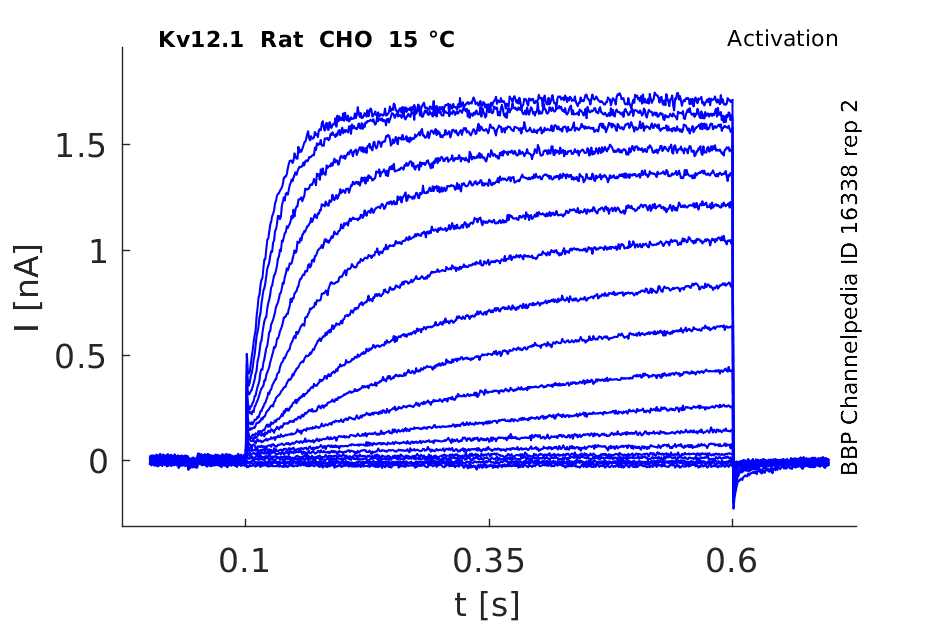
15 °Cshow 144 cells |
Click for details 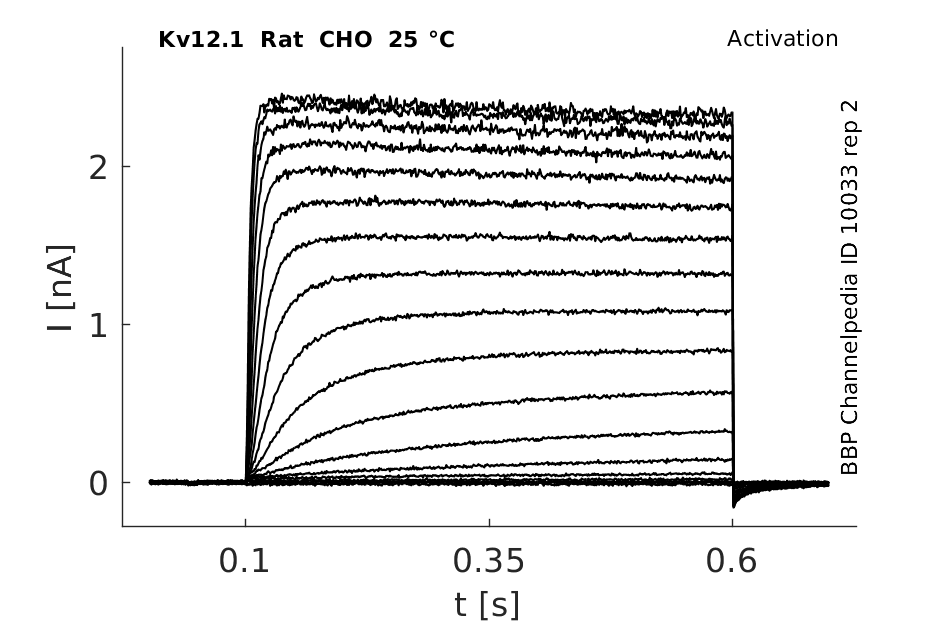
25 °Cshow 96 cells |
Click for details 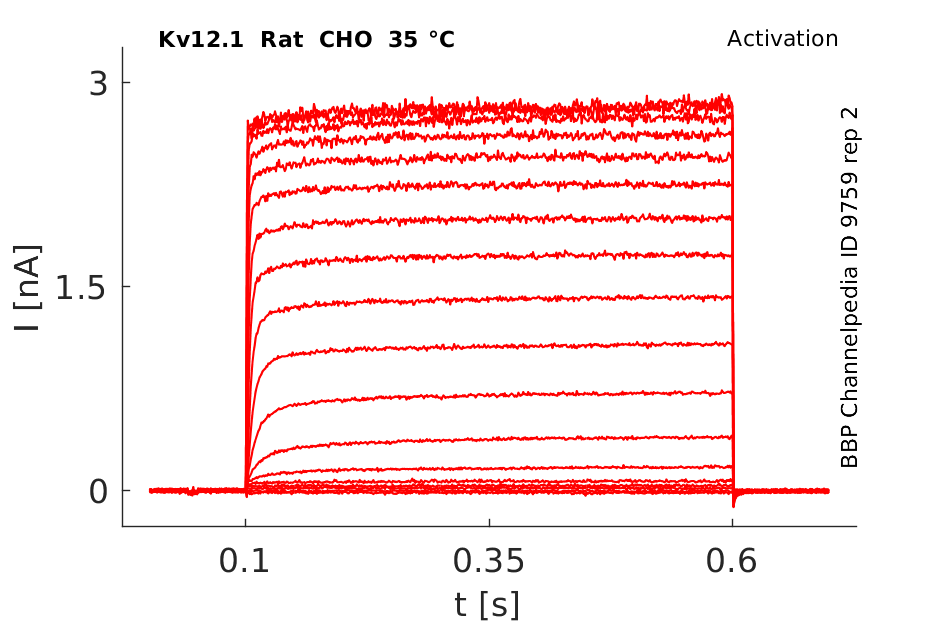
35 °Cshow 388 cells |
Transcript
| Species | NCBI accession | Length (nt) | |
|---|---|---|---|
| Human | NM_144633.3 | 5077 | |
| Mouse | NM_001031811.2 | 4305 | |
| Rat | NM_145095.2 | 3739 |
Protein Isoforms
Isoforms
Post-Translational Modifications
Visual Representation of Kv12.1 Structure
Methodology for visual representation of structure available here
Kv12.1 Crystal Structure

Kv12.1 predicted AlphaFold size
Methodology for AlphaFold size prediction and disclaimer are available here
rat Kv12.1 expressed in CHO cells
In rat elk1-transfected CHO cells, this protocol elicited slowly activating outward currents at test potentials positive to −70 mV. During the 2 s test pulses, RELK1-mediated outward currents did not inactivate. This behaviour was reminiscent of REAG-mediated outward currents. When test potentials were terminated by stepping back to the original holding potential of −120 mV, slowly deactivating tail currents were observed In Xenopus [809]
Kv12.1 recorded in Xenopus oocytes

Zebrafish Elk channel recorded in X oocytes

pH sensitivity of Kv12.1,Kv12.2 and Kv12.3

Expression of Kv12.1 in Human
Human KCNH8 is widely expressed in the human central nervous system and appears to overlap the distribution of human KCNH3 and KCNH4, which are also predominantly expressed in the nervous system. [808]
Kv12.1 was also identified in the sympathetic ganglia, testis, brain, colon, lung, uterus, pre-B cell leukemia (ESTs). [790], [809].
Kv12.1 in Rat Brain
Quantification by densitometry showed a very high level of Elk-1 mRNA expression in the granular layer of olfactory bulbs, the pyriform cortex, the dentate gyrus of the hippocampus, and the granular layer of the cerebellum. The labeling was relatively strong in the nucleus accumbens, the caudate–putamen, various nuclei of the thalamus, the CA layers in the hippocampus, the substantia nigra pars compacta, the pedonculopontine nucleus, and throughout the whole cerebral cortex. A low level of expression was observed in the lateral and medial septal nuclei, the substantia nigra pars reticulata (SNr), and the external segment of the globus pallidus (GPe). No significant hybridization signals were obtained in structures corresponding to white matter [1784]
Elk-1 protein detection in striatopallidal and striatonigral axon ter- minals. After unilateral kainic acid lesion of striatal cells, Elk-1 immunodetection was processed on sections corresponding to the external segment of the globus pallidus (GPe) and the substantia nigra pars reticulata (SNr) levels [1784]
Distribution of Kv12.1 in Rat Neurons
Multiple nuclear transcription factors including E-26-like protein 1 (Elk-1) have been found in neuronal dendrites, yet the functional significance of such localization has not yet been explained. Here we use a focal transfection procedure, 'phototransfection', to introduce Elk1 mRNA into specific regions of live, intact primary rat neurons. Introduction and translation of Elk1 mRNA in dendrites produced cell death, whereas introduction and translation of Elk1 mRNA in cell bodies did not produce cell death. Elk-1 translated in dendrites was transported to the nucleus, and cell death depended upon transcription, supporting the dendritic imprinting hypothesis and highlighting the importance of the dendritic environment on protein function [1783]
ELK1 for DNA Binding Modes
Eukaryotic transcription factors are grouped into families and, due to their similar DNA binding domains, often have the potential to bind to the same genomic regions. This can lead to redundancy at the level of DNA binding, and mechanisms are required to generate specific functional outcomes that enable distinct gene expression programmes to be controlled by a particular transcription factor. ELK1 uses different DNA binding modes to regulate functionally distinct classes of target genes [1779]
Prostate
Here, we investigated the expression and function of EPAC in human prostate tissues from patients undergoing radical prostatectomy. EPAC activation may reduce α1-adrenergic prostate contraction in the human prostate, although this effect is masked by cyclooxygenases and β-adrenoceptors. A main EPAC function in the human prostate may be the regulation of the transcription factor Elk1 [1755]
The ETS domain transcription factor ELK1 directs a critical component of growth signaling by the androgen receptor in prostate cancer cells [1758]
Stress stimulates differentiation of ligaments cells via ELK1 pathway
Cyclic tensile stress during physiological occlusal force enhances osteogenic differentiation of human periodontal ligament cells via ERK1/2-Elk1 MAPK pathway [1756]
ELK1 transcription factor linked to dysregulated striatal mu opioid receptor signalling
A characteristic feature of heroin abusers was decreased expression of MOR and extracellular regulated kinase signaling networks, concomitant with dysregulation of the downstream transcription factor ELK1. Striatal ELK1 in heroin abusers associated with the polymorphism rs2075572 in OPRM1 in a genotype dose-dependent manner and correlated with documented history of heroin use, an effect reproduced in an animal model that emphasizes a direct relationship between repeated heroin exposure and ELK1 dysregulation. A central role of ELK1 was evidenced by an unbiased whole transcriptome microarray that revealed ~20% of downregulated genes in human heroin abusers are ELK1 targets. ELK1 is a potential key transcriptional regulatory factor in striatal disturbances associated with heroin abuse and relevant to genetic mutation of OPRM1 [1757]
ELK1 controls cell Migration in Breast
Members of the ETS transcription factor family often target the same binding regions and hence have the potential to regulate the same genes and downstream biological processes. The ETS transcription factor ELK1 controls cell migration in breast epithelial cells through targeting a cohort of genes, independently from another family member GABPA, and therefore achieves biological specificity [1759]
Cancer
Ets1 and Elk1 transcription factors regulate cancerous inhibitor of protein phosphatase 2A expression in cervical and endometrial carcinoma cells [1778]
Inflammatory Bowel Diseases
Tumor necrosis factor (TNF-α) is a proinflammatory cytokine that plays a critical role in the pathogenesis of inflammatory bowel disease. TNF-α-induced increase in mouse intestinal permeability requires ERK1/2-dependent activation of Elk-1. These studies provide novel insight into the cellular and molecular processes that regulate the TNF-α-induced increase in intestinal epithelial tight junction permeability [1833]
Coexpression of KCNH8 and KCNH4
Distinct Elk channel subunits can coassemble with each other but that they likely do not coassemble with Eag and Erg family subunits. Coexpression of KCNH8 with dominant-negative KCNH8, KCNH3, and KCNH4 subunits led to suppression of the KCNH8 currents, suggesting that Elk channels can form heteromultimers. [808]
Nutlin-3
Nutlin-3 which occupies the p53 binding pocket in HDM2, has been reported to activate apoptosis through both the transcriptional activity-dependent and -independent programs of p53. Nutlin-3-activated ERK1/2 may stimulate the transcription of BCL2A1 via the activation of ELK1, and BCL2A1 expression may contribute to the inhibitory effect of ERK1/2 on nutlin-3-induced apoptosis, thereby constituting a negative feedback loop of p53-induced apoptosis [1776]
Katanin-p80
Katanin is an ATPase family member protein that participates in microtubule severing. We found that Elk1 activated KATNB1 promoter, and increased both mRNA and protein levels of katanin-p80 in SH-SY5Y cells. On the other hand, KCl treatment increasing SUMOylation decreased KATNB1 promoter activity. Since microtubule severing is an important cellular mechanism of which malfunctions result in serious diseases such as spastic paraplegia, Alzheimer's disease and cell cycle related disorders, identification of KATNB1 transcriptional regulation is crucial in understanding the coordination of microtubule severing activity by different proteins in the cells [1777]
SPG4
Mutations observed in SPG4 gene of hereditary spastic paraplegia patients have been shown to cause reduced spastin levels. In addition to mutations, transcriptional regulation of spastin gene expression may also affect spastin level. It has been identified the binding sites of Elk1 on the SPG4 promoter by EMSA. Over-expression of Elk1 showed that it repressed the SPG4 promoter and also decreased spastin protein level in SHSY-5Y cells [1780]
FADS2
A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1 [1781]
4-AP an Enhancer?
Interestingly, a RELK1 current increase (1.6-fold at 0 mV, n = 7) was observed after bath application of 10 mm 4-AP [809]
PIP2 inhibition of Kv12.1 (Elk1)voltage activation
Phosphatidylinositol 4,5-bisphosphate (PIP2) was shown to inhibit voltage activation of Elk1(KCNH8) but may also stabilize the open state. PIP2 inhibition of voltage activation requires the residues R347 in the S4-S5 linker and R479 near the S6 activation gate.[2085]
References
Zhang X
et al.
Divalent cations slow activation of EAG family K+ channels through direct binding to S4.
Biophys. J.,
2009
Jul
8
, 97 (110-20).
Shi W
et al.
Identification of two nervous system-specific members of the erg potassium channel gene family.
J. Neurosci.,
1997
Dec
15
, 17 (9423-32).
Warmke JW
et al.
A family of potassium channel genes related to eag in Drosophila and mammals.
Proc. Natl. Acad. Sci. U.S.A.,
1994
Apr
12
, 91 (3438-42).
Mayer SI
et al.
Signal transduction of pregnenolone sulfate in insulinoma cells: activation of Egr-1 expression involving TRPM3, voltage-gated calcium channels, ERK, and ternary complex factors.
J. Biol. Chem.,
2011
Mar
25
, 286 (10084-96).
Jiang L
et al.
Glycine-induced cytoprotection is mediated by ERK1/2 and AKT in renal cells with ATP depletion.
Eur. J. Cell Biol.,
2011
Apr
, 90 (333-41).
Wang W
et al.
The role of ERK-1/2 in the N/OFQ-induced inhibition of delayed rectifier potassium currents.
Biochem. Biophys. Res. Commun.,
2010
Apr
16
, 394 (1058-62).
Lee J
et al.
L-type calcium channel blockade on haloperidol-induced c-Fos expression in the striatum.
Neuroscience,
2007
Nov
9
, 149 (602-16).
Zou A
et al.
Distribution and functional properties of human KCNH8 (Elk1) potassium channels.
Am. J. Physiol., Cell Physiol.,
2003
Dec
, 285 (C1356-66).
Engeland B
et al.
Cloning and functional expression of rat ether-à-go-go-like K+ channel genes.
J. Physiol. (Lond.),
1998
Dec
15
, 513 ( Pt 3) (647-54).
Miyake A
et al.
New ether-à-go-go K(+) channel family members localized in human telencephalon.
J. Biol. Chem.,
1999
Aug
27
, 274 (25018-25).
Hennenberg M
et al.
The cAMP effector EPAC activates Elk1 transcription factor in prostate smooth muscle, and is a minor regulator of α1-adrenergic contraction.
J. Biomed. Sci.,
2013
, 20 (46).
Li L
et al.
Cyclic tensile stress during physiological occlusal force enhances osteogenic differentiation of human periodontal ligament cells via ERK1/2-Elk1 MAPK pathway.
DNA Cell Biol.,
2013
Sep
, 32 (488-97).
Sillivan SE
et al.
ELK1 transcription factor linked to dysregulated striatal mu opioid receptor signaling network and OPRM1 polymorphism in human heroin abusers.
Biol. Psychiatry,
2013
Oct
1
, 74 (511-9).
Patki M
et al.
The ETS domain transcription factor ELK1 directs a critical component of growth signaling by the androgen receptor in prostate cancer cells.
J. Biol. Chem.,
2013
Apr
19
, 288 (11047-65).
Odrowaz Z
et al.
The ETS transcription factors ELK1 and GABPA regulate different gene networks to control MCF10A breast epithelial cell migration.
PLoS ONE,
2012
, 7 (e49892).
Lee SY
et al.
Nutlin-3 induces BCL2A1 expression by activating ELK1 through the mitochondrial p53-ROS-ERK1/2 pathway.
Int. J. Oncol.,
2014
Aug
, 45 (675-82).
Selçuk E
et al.
Katanin-p80 gene promoter characterization and regulation via Elk1.
PLoS ONE,
2013
, 8 (e69423).
Pallai R
et al.
Ets1 and Elk1 transcription factors regulate cancerous inhibitor of protein phosphatase 2A expression in cervical and endometrial carcinoma cells.
Transcription,
2012 Nov-Dec
, 3 (323-35).
Odrowaz Z
et al.
ELK1 uses different DNA binding modes to regulate functionally distinct classes of target genes.
PLoS Genet.,
2012
, 8 (e1002694).
Canbaz D
et al.
SPG4 gene promoter regulation via Elk1 transcription factor.
J. Neurochem.,
2011
May
, 117 (724-34).
Lattka E
et al.
A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1.
J. Lipid Res.,
2010
Jan
, 51 (182-91).
Mo Y
et al.
Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA.
Nat. Struct. Biol.,
2000
Apr
, 7 (292-7).
Barrett LE
et al.
Region-directed phototransfection reveals the functional significance of a dendritically synthesized transcription factor.
Nat. Methods,
2006
Jun
, 3 (455-60).
Sgambato V
et al.
In vivo expression and regulation of Elk-1, a target of the extracellular-regulated kinase signaling pathway, in the adult rat brain.
J. Neurosci.,
1998
Jan
1
, 18 (214-26).
Brelidze TI
et al.
Structure of the carboxy-terminal region of a KCNH channel.
,
2012
Jan
9
, ().
Kazmierczak M
et al.
External pH modulates EAG superfamily K+ channels through EAG-specific acidic residues in the voltage sensor.
J. Gen. Physiol.,
2013
Jun
, 141 (721-35).
Al-Sadi R
et al.
TNF-α modulation of intestinal epithelial tight junction barrier is regulated by ERK1/2 activation of Elk-1.
Am. J. Pathol.,
2013
Dec
, 183 (1871-84).
Li X
et al.
Bimodal regulation of an Elk subfamily K+ channel by phosphatidylinositol 4,5-bisphosphate.
J. Gen. Physiol.,
2015
Nov
, 146 (357-74).
Contributors: Rajnish Ranjan, Nitin Khanna, Katherine Johnston
To cite this page: [Contributors] Channelpedia https://channelpedia.epfl.ch/wikipages/38/ , accessed on 2026 Mar 10