Cav3.1
Description: calcium channel, voltage-dependent, T type, alpha 1G subunit Gene: Cacna1g Alias: cacna1g, cav3.1, ca3.1, Ca(v)3.1
Cav3.1, encoded by the gene cacna1g, is a calcium channel, voltage-dependent, T type, alpha 1G subunit. It is highly expressed in the brain and heart, where it is responsible for long term potentiation and pacemaking, respectively. Mutations of the channel are the cause of developmental disorders and cardiopathies.
In humans, cacna1g, the gene which encodes Cav3.1, is composed of 38 exons located on chromosome 17 at position 21. (17q21.33). [2427]
Like most Cav channels, Cacna1g is subject to alternative splicing.
Some important exons, often involved in alternative splicing are [2481]:
- Exon 25: alternative splicing of the exon gives rise to either a1G-a or a1G-b. a1g-b alternates 59-splice donor site of exon 25 combined with the acceptor site on exon 27
- Exon 26: also known as insertion C which encodes for a section of the III-IV loop. It is generally only present alongside a1g-b
- Exon 14: also known as insertion e which encodes for a section of the II-III loop .
- Exon 34: also known as insertion f which encodes for a section of the COOH-terminal region.
- Exon 35: also known as insertion d which encodes for another section of the COOH-terminal region
| Species | NCBI accession | Length (nt) | |
|---|---|---|---|
| Human | NM_018896.5 | 8602 | |
| Mouse | NM_009783.3 | 8238 | |
| Rat | NM_001308302.1 | 7759 |
The human Cav3.1 protein is composed of 2377 amino acid (aa) and has a molecular weight of 262 Kda.
The CACNA1D gene undergoes extensive alternative splicing, leading to channels with subtle differences. The two variants, α1G-a and α1G-b, encode distinct intracellular III-IV loops. α1G-a is most similar to the canonical rat Cav3.1, while α1G-b has a shorter III-IV linker due to a 21-bp deletion.
These isoforms differ in expression: the α1G-a isoform, alone or combined with insertion e, is found in neuronal tissues, whereas α1G-b, mostly associated with insertion c, is detected in the fetal kidney. Additionally, α1G-b lacks the normally conserved phosphorylation sites. [2481]
Isoforms
A number of consensus phosphorylation sites for PKA and PKC were identified in Cav3.1, particularly in the intracellular regions, indicating that Cav3.1 may be subject to phosphorylation by these enzymes. [2481]
Phosphorylation is further evidenced by the activity of Rho-associated kinase (ROCK) on the channel. Activation of ROCK eversibly inhibited the peak current amplitudes of rat Ca(v)3.1 and Ca(v)3.3 channels without affecting the voltage dependence of activation or inactivation [2482]
Like most Cav channels, Cav3.1 is made up of a single protein composed of 4 homologous domains (DI-DIV). Each domain is made up of 6 transmembrane subunits (S1-S6) connected by extracellular loops. S1-4 form the voltage sensing domain (VSD) whereas the S5-S6 act as the selectivity filter and forms the pore module (PM). The S4 subunit of each domain contains a series of positively charged residues. [2483]
The structure of a Cav3.1 isoform was resolved via electron-microscopy, to a resolution of 3.3 angstrom. This splice variant of rat Cav3.1 has a deletion within the I–II linker, designated Cav3.1-Δ8b, and was shown to increase surface expression. Despite its differences with the canonical protein, Cryo-EM of Cav3.1-Δ8b allows for a detailed insight into the specific structural features of the protein. [2483]
The structural features of Cav3.1 are [2483]:
- Selectivity Filter (EEDD Motif): Cav3.1 has a unique selectivity filter composed of two Glutamate (Glu) and two Aspartate (Asp) residues, arranged as EEDD. This contrasts with the EEEE motif found in Cav1 and Cav2 channels, contributing to its distinctive calcium ion selectivity.
- Disulfide Bond: A unique disulfide bond between Cys104 and Cys889 is present in Cav3.1, stabilizing the interaction between the voltage-sensing domain (VSD) I and the pore domain. This bond is specific to T-type channels like Cav3.1 and is involved in redox modulation.
- Incompatibility with Auxiliary Subunits: Cav3.1 lacks the structural elements needed for binding with auxiliary subunits like α2δ, which are present in high-voltage-activated (HVA) channels. Specific extracellular loops (ECLI and ECLIII) in Cav3.1 would physically clash with these subunits, preventing their association.
- Pore Domain Structure: The S5 and S6 helices form a pore domain that is characterized by a smaller constriction site compared to L-type channels. This structure is crucial for the specific ion conductance properties of Cav3.1.
These features collectively distinguish Cav3.1 from other voltage-gated calcium channels and contribute to its specific physiological functions.
Cav3.1 predicted AlphaFold size
Methodology for AlphaFold size prediction and disclaimer are available here
Cav3.1 channels generate significant calcium currents by allowing entry of calcium into the cell in response to action potentials. [2484]
CaV3.1, CaV3.2, and CaV3.3 are characterized by their activation at low membrane potentials, close to the resting membrane potential, and transient single-channel conductance and are thus designated as the low-voltage-activated (LVA) or T-type channels. This is in contrast to Cav1 and Cav2, which are high-voltage-activated and conduct larger, long-lasting conductance. [2485] [2483]
The low voltage threshold for activation of T-type channels drives their opening in response to relatively small positive changes in membrane potential. Further, an overlap in the membrane potentials at which T-type channels open, close, and inactivate gives them a particular property known as a “window current” whereby a basal inward flux of calcium ions can occur near the resting potential [2484]
Cav3.1 also has its own particular gating properties [2484] [2486]:
- Activation and Inactivation Kinetics: Cav3.1 channels have the fastest activation and inactivation kinetics among the T-type channels .
- Voltage Dependence: Cav3.1 channels open and close at similar membrane potentials as Cav3.2 channels, but at more hyperpolarized potentials compared to Cav3.3.
- Deactivation Kinetics: Cav3.1 channels deactivate (close) faster than Cav3.2 channels but slower than Cav3.3 channels.
- Recovery from Inactivation: Cav3.1 channels recover from inactivation more quickly than Cav3.2 and Cav3.3 channels.
Single channel unitary conductance
The single channel unitary conductance of Cav3.1 was at around 7 pS. [2481]
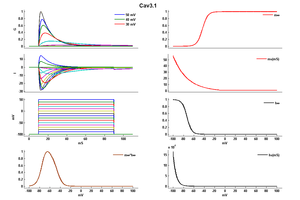
Model
Model Cav3.1 (ID=41)
| Animal | CH | |
| CellType | CHO | |
| Age | 0 Days | |
| Temperature | 0.0°C | |
| Reversal | 30.0 mV | |
| Ion | Ca + | |
| Ligand ion | ||
| Reference | [103] Achraf Traboulsie et. al; J. Physiol. (Lond.) 2007 Jan 1 | |
| mpower | 1.0 | |
| m Inf | 1 /(1+exp((v-(-42.921064))/-5.163208)) | |
| m Tau | -0.855809 + (1.493527 * exp(-v/27.414182)) If v lt -10 | |
| m Tau | 1.0 If v gteq -10 | |
| hpower | 1.0 | |
| h Inf | 1 /(1+exp((v-(-72.907420))/4.575763)) | |
| h Tau | 9.987873 + (0.002883 * exp(-v/5.598574)) | |
Tissues & Cellular
Cav3.1 can be found in different locations of the body. However, it’s predominant highly expressed in the brain, particularly in the thalamus, cerebellum (Purkinje neurons ), substantia nigra, and frontal and occipital lobes [2481] [2487]
Cav3.1 is also present at significant levels in the heart, including the sinoatrial node (SAN) and atrioventricular node (AVN), as well as in muscle sarcoma, the colon, lungs, breast, kidney, and reproductive organs such as the ovaries, placenta, testes, and prostate. [2399] [2422] [2481]
Developmental
Cav3.1 expression if thought tp be developmentally regulated. Though the channels expression is comparable in adult and fetal brains, it is more abundant in fetal peripheral tissues like the heart, kidneys, and lungs. This expression corroborates with fetal heart electrophysiological data, as T-type currents are preferentially recorded in embryonic heart cells. [2481]
There is little research on the suc-cellular location of Cav3.1 and location is likely dependent on the type of cell where the channel is present.
In retina nerve cells, Cav3.1 is localized to the somatodendritic compartment and proximal axon [2488]
Cav3.1’s role is to allow the entry of Ca2+ into the cell in a voltage dependent manner. Depending on its expressed location, the channel fulfills a number of function
Neuronal signalling
Cav3.1 is responsible for postsynaptic calcium signaling and long term potentiation. Among the T-type channels, CaV3.1 (α1G) channels produce larger, transient calcium currents that are essential for initiating action potentials. [2486] [2415]
Depending on which nerve cells and brain region they are present, the channels can have further functions. In neurons of the hypothalamic paraventricular nucleus, Cav3.1 is the major subtype that mediates T-type calcium channel-dependent low-threshold spikes (LTSs) [2489] In the thalamocortical relay neurons, CaV3.1 channels contribute significantly to burst firing in neurons and mediate low-threshold spikes essential for certain types of rhythmic activity, such as those observed in sleep and epilepsy. [2486]. KO Cav3.1 mice display no burst firing activity, were less prone to developing tonic seizures but did display some issues with motor performance and cerebellar learning [2487]
Sleep
Impaired Cav3.1 channels were shown to decrease sleep duration [2490] KO mice lacking the T-channel Cav3.1 subunit demonstrated that the activation of T-channels during wake-like states is a major determinant for single and multiple spike occurrence during tonic firing and for the robustness of the thalamocortical transfer of sensory inputs during sleep [2491]
Pacemaking
In the heart, Cav3.1 contributed to heart rate genesis by ensuring rep [2492] Disruption of channel abolishes the T-type calcium current in the SAN and the atrioventricular node. Inactivation of Cav3.1 lead to a significant slowing of in vivo heart rate, prolonged the SAN recovery time, and slowed pacemaker activity of individual SAN cells through a reduction of the slope of the diastolic depolarization [530]
Pain
Cav3.1 is not responsible for pain signalling as the channel is not expressed in the sensory ganglia and KO models reveled normal peripheral pain detection [2485]
Channelopathies
Given the importance of Cav3.1’s role through the body, mutations to coding gene or disruption to the protein are responsible for a number of pathologies
Developmental disorders
Given its important and predominant expression in the brain, aberrant Cav3.1 function leads to a number of cognitive impairment. These include [2487]:
- Late-onset cerebellar ataxia ADCA/SCA42: Hereditary cerebellar ataxias are rare neurodegenerative disorders, characterized by a cerebellar syndromewith or without other neurological symptomsCACNA1G was linked to an autosomal dominant cerebellar ataxia (ADCA)
- Childhood cerebellar atrophy: is a devastating infantile neurodevelopmental disorder, with severe motor and cognitive impairments, cerebellar atrophy, and variable features including facial dysmorphism, digital anomalies, microcephaly, hirsutism, and epilepsy.
- Autism spectrum disorder: CACNA1G was identified as a candidate gene for autism spectrum disorder (ASD)
- Epilepsy: Idiopathic generalized epilepsies (IGEs), Juvenile myoclonic epilepsy, severe developmental and epileptic encephalopathies (DEEs), genetic generalized epilepsy (GGE) [2493]
- Parkinsons: Cav3.1 is the predominant form of T‐type channels in the dopaminergic neurons of the Substantia nigra pars compacta. SN is thought to play an emerging role in Parkinsons disorder. Cav3.1 TTCCs stabilize SN DA pacemaker frequency and its precision in an age‐dependent manner and therefore may have a hand in the development of parkinson [2494]
Cardiopathies
Cav3.1 is cruchal for the proper function of pacemaker cells in the sinoatrial node of the heart. These channels help regulate the heart's rhythm by contributing to the diastolic. Cav3.1 is a pathophysiological target for [2495]: Sinus Node Dysfunction (SND): Mutations in Cav3.1 channels are linked to sinus node dysfunction, a condition where the heart's natural pacemaker fails to function properly. This can lead to irregular heart rhythms (arrhythmias) or bradycardia (slow heart rate). Heart Block: Cav3.1 dysfunction can also contribute to atrioventricular (AV) block, where the electrical signals between the heart's chambers are delayed or blocked, potentially leading to severe arrhythmias [2495]
Cancer
Aberrant expression of cacna1g is common in a broad range of cancers. Cav3.1 is the top 1% of overexpressed genes in synovial sarcoma and prostate carcinoma but it is also highly expressed in other tumors such as colorectal, uterine, prostate, and breast cancer. Depending on the type, cacna1g can also be under-expressed in breast cancer [2422] [2440]
The CpG island methylator phenotype (CIMP or CIMP-high) with extensive promoter methylation is a distinct phenotype in cancer, particularly colorectal cancer. A high volume of evidence has shown that high methylation of Cacna1g promoter is an epigenetic marker for tumours.[2496] [2497] [2498] [2499] [2500] [2501]
Neonatal methylation markers on Cacna1g were also significantly associated with birth weight. [2502]
Endocrinopathies
Mutation in CACNA1D were found to cause aldosterone producing adenoma (APA) and familial hyperaldosteronism (FH)
Interestingly, Cav3.1 channels of function in the absence of auxiliary subunits as they lack the proper binding sites for proper interaction (see)Structure [2483] Indeed similarly, the G protein binding sites have not been identified nor Ca2+ and calmodulin binding domains involved in calcium dependent inactivation [2481]
Cav3.1 is sensitive to ions, particularly Zn2+. ZN2+ inhibition is associated with a shift to more negative membrane potentials of both Cav3.1 steady-state inactivation and steady-state activation currents. There are also changes in kinetics, especially a significant slowing of the inactivation kinetics. Application of Zn21 results significant reduction in current.[103]
Another notable inhibitor of T-type channels is Mibefradil. [2503]
CACHD1 is a protein highly expressed in the male mammalian CNS with similar distribution patterns as Cav3. subunits. CACHD1 was shown to increase cell-surface localization of CaV3.1, and caused a significant increase in peak current density and corresponding increases in maximal conductance, resulting in increased open probability of the channel and neuronal excitability. [2504]
Estrogen increases the expression of the Cav3.1 in certain brain regions, notablyin the hypothalamus and pituitary. The hormone also enhances the activity of these channels in neurons, leading to increased calcium currents and more burst firing. These modulations suggest that estrogen boosts the function of T-type calcium channels, which in turn enhances neuron firing and neurotransmitter release in the brain. [2505]
References
Traboulsie A
et al.
Subunit-specific modulation of T-type calcium channels by zinc.
J. Physiol. (Lond.),
2007
Jan
1
, 578 (159-71).
Hansen JP
et al.
Calcium channel gamma6 subunits are unique modulators of low voltage-activated (Cav3.1) calcium current.
J. Mol. Cell. Cardiol.,
2004
Dec
, 37 (1147-58).
Niwa N
et al.
Cav3.2 subunit underlies the functional T-type Ca2+ channel in murine hearts during the embryonic period.
Am. J. Physiol. Heart Circ. Physiol.,
2004
Jun
, 286 (H2257-63).
Lacinova L
et al.
Modulation of gating currents of the Ca(v)3.1 calcium channel by alpha 2 delta 2 and gamma 5 subunits.
Arch. Biochem. Biophys.,
2004
May
15
, 425 (207-13).
Emerick MC
et al.
Profiling the array of Ca(v)3.1 variants from the human T-type calcium channel gene CACNA1G: alternative structures, developmental expression, and biophysical variations.
Proteins,
2006
Aug
1
, 64 (320-42).
Kovacs K
et al.
Subcellular distribution of low-voltage activated T-type Ca2+ channel subunits (Ca(v)3.1 and Ca(v)3.3) in reticular thalamic neurons of the cat.
J. Neurosci. Res.,
2010
Feb
1
, 88 (448-60).
Catterall WA
Structure and regulation of voltage-gated Ca2+ channels.
Annu. Rev. Cell Dev. Biol.,
2000
, 16 (521-55).
Perez-Reyes E
Molecular physiology of low-voltage-activated t-type calcium channels.
Physiol. Rev.,
2003
Jan
, 83 (117-61).
Hagiwara N
et al.
Contribution of two types of calcium currents to the pacemaker potentials of rabbit sino-atrial node cells.
J. Physiol. (Lond.),
1988
Jan
, 395 (233-53).
Mangoni ME
et al.
Bradycardia and slowing of the atrioventricular conduction in mice lacking CaV3.1/alpha1G T-type calcium channels.
Circ. Res.,
2006
Jun
9
, 98 (1422-30).
Huguenard JR
Low-threshold calcium currents in central nervous system neurons.
Annu. Rev. Physiol.,
1996
, 58 (329-48).
Anderson MP
et al.
Thalamic Cav3.1 T-type Ca2+ channel plays a crucial role in stabilizing sleep.
Proc. Natl. Acad. Sci. U.S.A.,
2005
Feb
1
, 102 (1743-8).
Chen XL
et al.
A role for T-type Ca2+ channels in the synergistic control of aldosterone production by ANG II and K+.
Am. J. Physiol.,
1999
May
, 276 (F674-83).
Arnoult C
et al.
Activation of mouse sperm T-type Ca2+ channels by adhesion to the egg zona pellucida.
Proc. Natl. Acad. Sci. U.S.A.,
1996
Nov
12
, 93 (13004-9).
Tsakiridou E
et al.
Selective increase in T-type calcium conductance of reticular thalamic neurons in a rat model of absence epilepsy.
J. Neurosci.,
1995
Apr
, 15 (3110-7).
Todorovic SM
et al.
Pharmacological properties of T-type Ca2+ current in adult rat sensory neurons: effects of anticonvulsant and anesthetic agents.
J. Neurophysiol.,
1998
Jan
, 79 (240-52).
Nuss HB
et al.
T-type Ca2+ current is expressed in hypertrophied adult feline left ventricular myocytes.
Circ. Res.,
1993
Oct
, 73 (777-82).
Larsen JK
et al.
Quantitative analysis of the expression and distribution of calcium channel alpha 1 subunit mRNA in the atria and ventricles of the rat heart.
J. Mol. Cell. Cardiol.,
2002
May
, 34 (519-32).
Marionneau C
et al.
Specific pattern of ionic channel gene expression associated with pacemaker activity in the mouse heart.
J. Physiol. (Lond.),
2005
Jan
1
, 562 (223-34).
Kessi M
et al.
Calcium channelopathies and intellectual disability: a systematic review.
Orphanet J Rare Dis, 2021May13, 16 (219).
Wang CY
et al.
Meta-Analysis of Public Microarray Datasets Reveals Voltage-Gated Calcium Gene Signatures in Clinical Cancer Patients.
PLoS ONE,
2015
, 10 (e0125766).
Rajakulendran S
et al.
Neuronal P/Q-type calcium channel dysfunction in inherited disorders of the CNS.
,
2012
Jan
17
, ().
Phan NN
et al.
Voltage-gated calcium channels: Novel targets for cancer therapy.
Oncol Lett, 2017Aug, 14 (2059-2074).
Monteil A
et al.
Molecular and functional properties of the human alpha(1G) subunit that forms T-type calcium channels.
J. Biol. Chem.,
2000
Mar
3
, 275 (6090-100).
Iftinca M
et al.
Regulation of T-type calcium channels by Rho-associated kinase.
Nat. Neurosci.,
2007
Jul
, 10 (854-60).
Zhao Y
et al.
Cryo-EM structures of apo and antagonist-bound human Cav3.1.
Nature, 2019Dec, 576 (492-497).
Cain SM
et al.
Contributions of T-type calcium channel isoforms to neuronal firing.
Channels (Austin),
2010 Nov-Dec
, 4 (475-82).
Bourinet E
et al.
Silencing of the Cav3.2 T-type calcium channel gene in sensory neurons demonstrates its major role in nociception.
EMBO J.,
2005
Jan
26
, 24 (315-24).
Chemin J
et al.
Specific contribution of human T-type calcium channel isotypes (alpha(1G), alpha(1H) and alpha(1I)) to neuronal excitability.
J. Physiol. (Lond.),
2002
Apr
1
, 540 (3-14).
Lory P
et al.
Neuronal Cav3 channelopathies: recent progress and perspectives.
Pflugers Arch, 2020Jul, 472 (831-844).
Puthussery T
et al.
NaV1.1 channels in axon initial segments of bipolar cells augment input to magnocellular visual pathways in the primate retina.
J. Neurosci.,
2013
Oct
9
, 33 (16045-59).
Zhang C
et al.
17Beta-estradiol regulation of T-type calcium channels in gonadotropin-releasing hormone neurons.
J. Neurosci.,
2009
Aug
26
, 29 (10552-62).
Tatsuki F
et al.
Involvement of Ca(2+)-Dependent Hyperpolarization in Sleep Duration in Mammals.
Neuron, 2016Apr06, 90 (70-85).
Deleuze C
et al.
T-type calcium channels consolidate tonic action potential output of thalamic neurons to neocortex.
J. Neurosci.,
2012
Aug
29
, 32 (12228-36).
Li Y
et al.
Increasing T-type calcium channel activity by β-adrenergic stimulation contributes to β-adrenergic regulation of heart rates.
J Physiol, 2018Apr01, 596 (1137-1151).
Ultra-Rare Genetic Variation in the Epilepsies: A Whole-Exome Sequencing Study of 17,606 Individuals.
Am J Hum Genet, 2019Aug01, 105 (267-282).
Duda J
et al.
Converging roles of ion channels, calcium, metabolic stress, and activity-pattern of substantia nigra dopaminergic neurons in health and Parkinson's disease.
J. Neurochem.,
2016
Feb
10
, ().
Torrente AG
et al.
Channelopathies of voltage-gated L-type Cav1.3/α1D and T-type Cav3.1/α1G Ca2+ channels in dysfunction of heart automaticity.
Pflugers Arch, 2020Jul, 472 (817-830).
Ogino S
et al.
Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample.
J Mol Diagn,
2007
Jul
, 9 (305-14).
Ogino S
et al.
Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype.
Clin Cancer Res, 2009Oct15, 15 (6412-20).
Ogino S
et al.
CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer.
Gut, 2009Jan, 58 (90-6).
Nosho K
et al.
Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample.
PLoS One, 2008, 3 (e3698).
Ogino S
et al.
CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations.
J Mol Diagn, 2006Nov, 8 (582-8).
Lin X
et al.
Developmental pathways to adiposity begin before birth and are influenced by genotype, prenatal environment and epigenome.
BMC Med, 2017Mar07, 15 (50).
Lacinova L
Pharmacology of recombinant low-voltage activated calcium channels.
,
2004
Apr
, 3 (105-11).
Cottrell GS
et al.
CACHD1 is an α2δ-Like Protein That Modulates CaV3 Voltage-Gated Calcium Channel Activity.
J Neurosci, 2018Oct24, 38 (9186-9201).
Qiu J
et al.
Estrogen upregulates T-type calcium channels in the hypothalamus and pituitary.
J. Neurosci.,
2006
Oct
25
, 26 (11072-82).
Contributors: Rajnish Ranjan, Michael Schartner
To cite this page: [Contributors] Channelpedia https://channelpedia.epfl.ch/wikipages/85/ , accessed on 2026 Mar 01