Kir3.1
Description: potassium inwardly-rectifying channel, subfamily J, member 3 Gene: Kcnj3 Alias: Kir3.1, GIRK1, Kcnf3, Kcnj3
KCNJ3 (also known as KGA; GIRK1; KIR3.1) encodes the integral membrane protein Kir3.1, a potassium inwardly-rectifying channel, subfamily J, member 3. The encoded protein, which has a greater tendency to allow potassium to flow into a cell rather than out of a cell, is controlled by G-proteins and plays an important role in regulating heartbeat. It associates with three other G-protein-activated potassium channels to form a heteromultimeric pore-forming complex.
http://www.ncbi.nlm.nih.gov/gene/3760
Experimental data
Rat Kir3.1 gene in CHO host cells |
||
|
Click for details 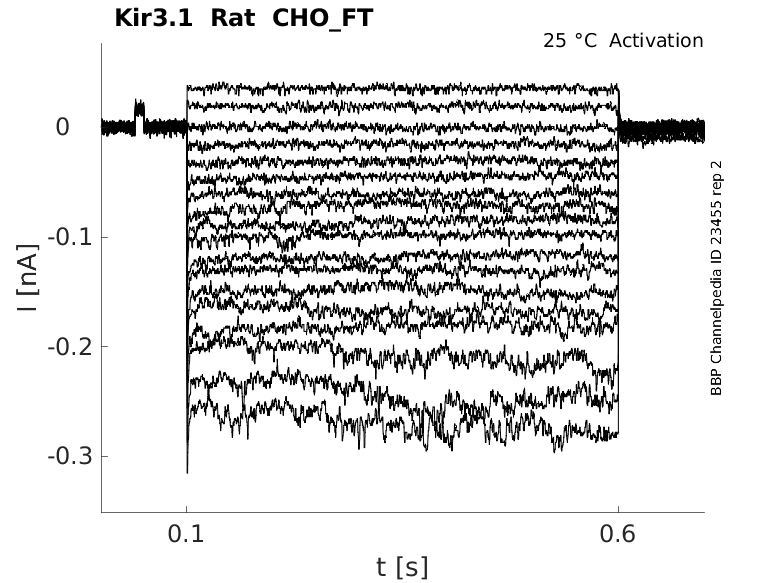
25 °Cshow 48 cells |
Click for details 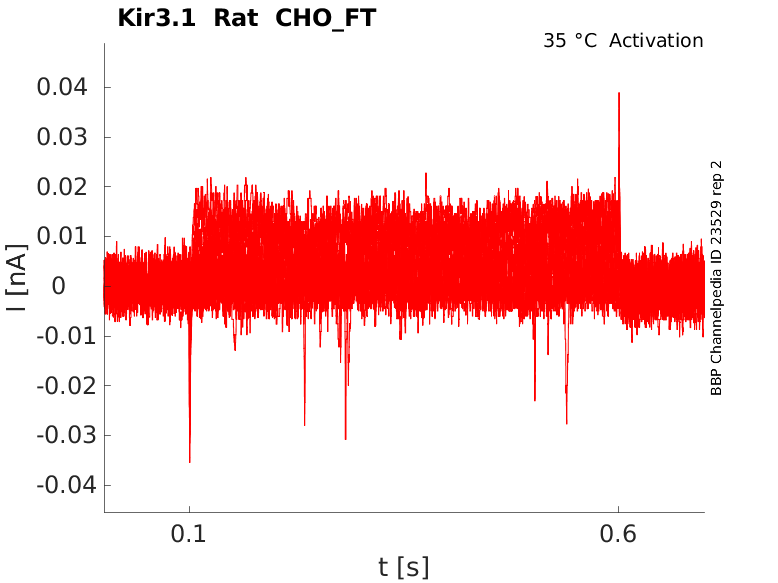
35 °Cshow 23 cells |
|
The Kir3.1 gene contains three exons separated by two introns, and its total length exceeds 45 kb. The two transmembrane domains, pore region, and part of the putative carboxyl terminus are encoded by exon 1, whereas the remainder of the tail is encoded by exons 2 and 3. The mRNA transcription initiation site was established, and the first 1520 bp upstream were sequenced; this region lacked a traditional TATA or CAAT box, but contained a GC-rich region as well as various putative transcription factor-binding elements. The 1520 bp upstream and 84 bp downstream of the transcription initiation site were tested for promoter activity in GH4-C1 cells. This sequence of 1604 bp contains a number of fragments that either stimulate or repress transcription, as tested by transient expression of various Kir3.1 promoter/luciferase fusion gene constructs in GH4-C1 cells. Schoots 1997 [966]
Transcript
| Species | NCBI accession | Length (nt) | |
|---|---|---|---|
| Human | NM_002239.4 | 4628 | |
| Mouse | NM_008426.2 | 5232 | |
| Rat | NM_031610.3 | 1827 |
Protein Isoforms
Isoforms
Post-Translational Modifications
Structure
Kir3.1 predicted AlphaFold size
Methodology for AlphaFold size prediction and disclaimer are available here
Single Channel Recording of GIRK1 in CHO cells

Kir3.1 is expressed in cells of different lineage, i.e., cardiac atrial myocytes as well as various neuronal cell types. Currently no human cell line is known to express the Kir3.1 channel. Schoots 1997 [966]
G protein-gated inwardly rectifying potassium (GIRK or Kir3) channel activity is important for regulating excitability in the heart and brain (Stanfield et al. 2002 [957]).
Kir3.1 channel is involved in the TLR4-mediated (Toll-like receptor in immune system) signal at an early event by facilitating the recruitment of TLR4 into lipid raft. [961]
G-protein Interaction
Kir3 channels are activated following stimulation of G protein-coupled receptors (GPCRs) that use the Gi/o family of G proteins. Stimulation of the GPCR promotes exchange of GDP for GTP on the Gα subunit which, in turn, leads to activation of the Gα subunit and the Gβγ dimer. Gβγ dimers bind to and activate Kir3 channels (Reuveny et al. 1994 [968]; Wickman et al. 1994 [969]; Huang et al. 1995 [970]). Gα subunits are required for terminating Kir3 activation. The intrinsic GTPase activity of the Gα subunit hydrolyses GTP, leading to inactivation of the Gβγ dimer. Regulator of G protein signalling (RGS) proteins accelerate the GTPase activity of Gα subunits (GAP), leading to faster activation and deactivation of Kir3 channels (Doupnik et al. 1997 [971]). (From Fowler [965])
Heterotetramers with other kir3
The Kir3 family consists of the Kir3.1, Kir3.2, Kir3.3, and Kir3.4 subunits, and the majority of functional Kir3 channels are believed to exist as heterotetram- ers containing the Kir3.1 subunit, although some studies report on functional Kir3.2 homomers and Kir3.2/3.3 combinations (Wischmeyer et al., 1997; Inanobe et al., 1999; Jelacic et al., 2000).
Constitutive and agonist-induced interactions between Kir3 channels and various Gβγ combinations in living cells are reported by Riven [967].
Gβ1-4 can interact with Kir3.1 in the absence of Kir3.4. Gβ5 does not directly interact with the channel but can still be co-immunoprecipated as part of a larger complex.[963]
References
Kim D
et al.
ATP-dependent regulation of a G protein-coupled K+ channel (GIRK1/GIRK4) expressed in oocytes.
Am. J. Physiol.,
1997
Jan
, 272 (H195-206).
Stanfield PR
et al.
Constitutively active and G-protein coupled inward rectifier K+ channels: Kir2.0 and Kir3.0.
Rev. Physiol. Biochem. Pharmacol.,
2002
, 145 (47-179).
Jo HY
et al.
Kir3.1 channel is functionally involved in TLR4-mediated signaling.
Biochem. Biophys. Res. Commun.,
2011
Apr
22
, 407 (687-91).
Styer AM
et al.
G protein {beta}{gamma} gating confers volatile anesthetic inhibition to Kir3 channels.
J. Biol. Chem.,
2010
Dec
31
, 285 (41290-9).
Robitaille M
et al.
Intracellular trafficking and assembly of specific Kir3 channel/G protein complexes.
Cell. Signal.,
2009
Apr
, 21 (488-501).
Brevet M
et al.
Expression of K+ channels in normal and cancerous human breast.
Histol. Histopathol.,
2008
Aug
, 23 (965-72).
Fowler CE
et al.
Evidence for association of GABA(B) receptors with Kir3 channels and regulators of G protein signalling (RGS4) proteins.
J. Physiol. (Lond.),
2007
Apr
1
, 580 (51-65).
Schoots O
et al.
Genomic organization and promoter analysis of the human G-protein-coupled K+ channel Kir3.1 (KCNJ3/HGIRK1).
Genomics,
1997
Feb
1
, 39 (279-88).
Riven I
et al.
GIRK channel activation involves a local rearrangement of a preformed G protein channel complex.
Neuron,
2006
Sep
7
, 51 (561-73).
Reuveny E
et al.
Activation of the cloned muscarinic potassium channel by G protein beta gamma subunits.
Nature,
1994
Jul
14
, 370 (143-6).
Wickman KD
et al.
Recombinant G-protein beta gamma-subunits activate the muscarinic-gated atrial potassium channel.
Nature,
1994
Mar
17
, 368 (255-7).
Huang CL
et al.
Evidence that direct binding of G beta gamma to the GIRK1 G protein-gated inwardly rectifying K+ channel is important for channel activation.
Neuron,
1995
Nov
, 15 (1133-43).
Doupnik CA
et al.
RGS proteins reconstitute the rapid gating kinetics of gbetagamma-activated inwardly rectifying K+ channels.
Proc. Natl. Acad. Sci. U.S.A.,
1997
Sep
16
, 94 (10461-6).
Delling M
et al.
The neural cell adhesion molecule regulates cell-surface delivery of G-protein-activated inwardly rectifying potassium channels via lipid rafts.
J. Neurosci.,
2002
Aug
15
, 22 (7154-64).
Velimirovic BM
et al.
The K+ channel inward rectifier subunits form a channel similar to neuronal G protein-gated K+ channel.
FEBS Lett.,
1996
Jan
22
, 379 (31-7).
Credits
To cite this page: [Contributors] Channelpedia https://channelpedia.epfl.ch/wikipages/46/ , accessed on 2026 Feb 19